Hong An
SwarmThinkers: Learning Physically Consistent Atomic KMC Transitions at Scale
May 26, 2025Abstract:Can a scientific simulation system be physically consistent, interpretable by design, and scalable across regimes--all at once? Despite decades of progress, this trifecta remains elusive. Classical methods like Kinetic Monte Carlo ensure thermodynamic accuracy but scale poorly; learning-based methods offer efficiency but often sacrifice physical consistency and interpretability. We present SwarmThinkers, a reinforcement learning framework that recasts atomic-scale simulation as a physically grounded swarm intelligence system. Each diffusing particle is modeled as a local decision-making agent that selects transitions via a shared policy network trained under thermodynamic constraints. A reweighting mechanism fuses learned preferences with transition rates, preserving statistical fidelity while enabling interpretable, step-wise decision making. Training follows a centralized-training, decentralized-execution paradigm, allowing the policy to generalize across system sizes, concentrations, and temperatures without retraining. On a benchmark simulating radiation-induced Fe-Cu alloy precipitation, SwarmThinkers is the first system to achieve full-scale, physically consistent simulation on a single A100 GPU, previously attainable only via OpenKMC on a supercomputer. It delivers up to 4963x (3185x on average) faster computation with 485x lower memory usage. By treating particles as decision-makers, not passive samplers, SwarmThinkers marks a paradigm shift in scientific simulation--one that unifies physical consistency, interpretability, and scalability through agent-driven intelligence.
Predictive Accuracy-Based Active Learning for Medical Image Segmentation
May 01, 2024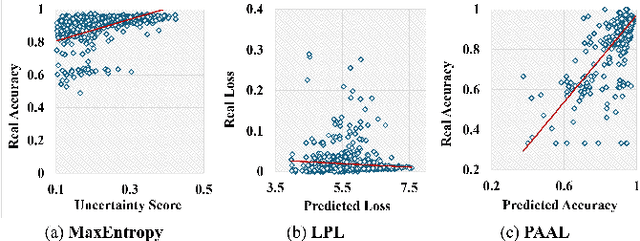
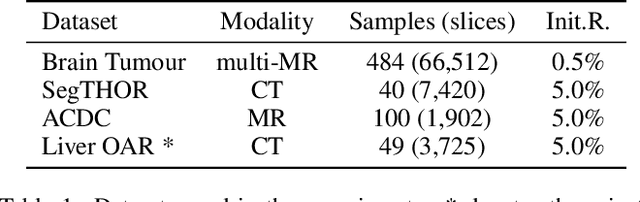
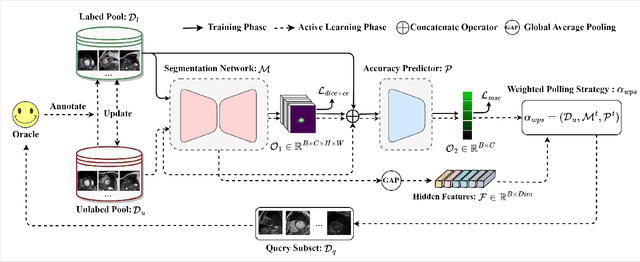
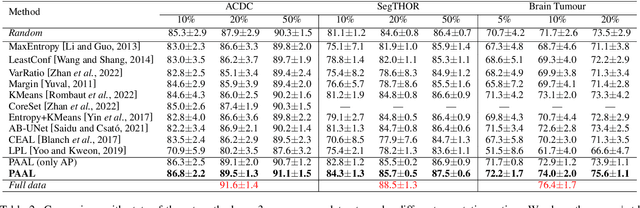
Abstract:Active learning is considered a viable solution to alleviate the contradiction between the high dependency of deep learning-based segmentation methods on annotated data and the expensive pixel-level annotation cost of medical images. However, most existing methods suffer from unreliable uncertainty assessment and the struggle to balance diversity and informativeness, leading to poor performance in segmentation tasks. In response, we propose an efficient Predictive Accuracy-based Active Learning (PAAL) method for medical image segmentation, first introducing predictive accuracy to define uncertainty. Specifically, PAAL mainly consists of an Accuracy Predictor (AP) and a Weighted Polling Strategy (WPS). The former is an attached learnable module that can accurately predict the segmentation accuracy of unlabeled samples relative to the target model with the predicted posterior probability. The latter provides an efficient hybrid querying scheme by combining predicted accuracy and feature representation, aiming to ensure the uncertainty and diversity of the acquired samples. Extensive experiment results on multiple datasets demonstrate the superiority of PAAL. PAAL achieves comparable accuracy to fully annotated data while reducing annotation costs by approximately 50% to 80%, showcasing significant potential in clinical applications. The code is available at https://github.com/shijun18/PAAL-MedSeg.
Pruner: An Efficient Cross-Platform Tensor Compiler with Dual Awareness
Feb 04, 2024Abstract:Tensor program optimization on Deep Learning Accelerators (DLAs) is critical for efficient model deployment. Although search-based Deep Learning Compilers (DLCs) have achieved significant performance gains compared to manual methods, they still suffer from the persistent challenges of low search efficiency and poor cross-platform adaptability. In this paper, we propose $\textbf{Pruner}$, following hardware/software co-design principles to hierarchically boost tensor program optimization. Pruner comprises two primary components: a Parameterized Static Analyzer ($\textbf{PSA}$) and a Pattern-aware Cost Model ($\textbf{PaCM}$). The former serves as a hardware-aware and formulaic performance analysis tool, guiding the pruning of the search space, while the latter enables the performance prediction of tensor programs according to the critical data-flow patterns. Furthermore, to ensure effective cross-platform adaptation, we design a Momentum Transfer Learning ($\textbf{MTL}$) strategy using a Siamese network, which establishes a bidirectional feedback mechanism to improve the robustness of the pre-trained cost model. The extensive experimental results demonstrate the effectiveness and advancement of the proposed Pruner in various tensor program tuning tasks across both online and offline scenarios, with low resource overhead. The code is available at https://github.com/qiaolian9/Pruner.
H-DenseFormer: An Efficient Hybrid Densely Connected Transformer for Multimodal Tumor Segmentation
Jul 04, 2023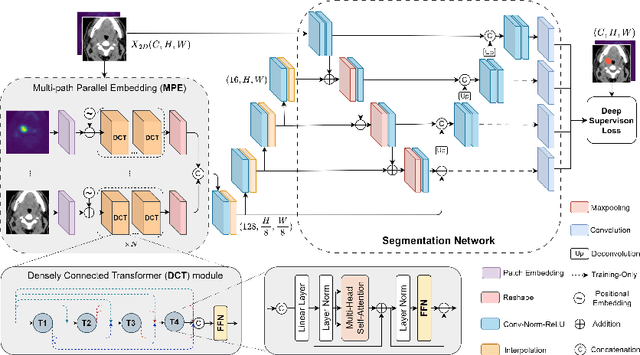
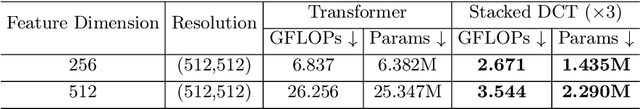
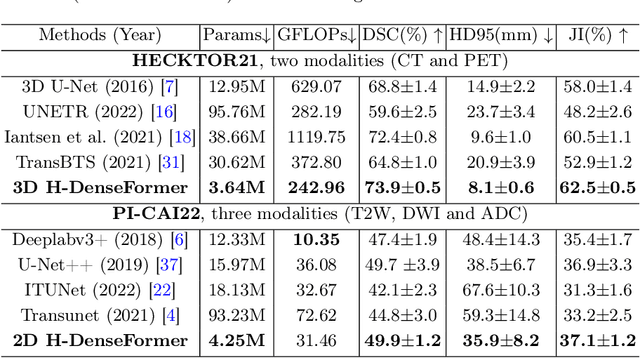
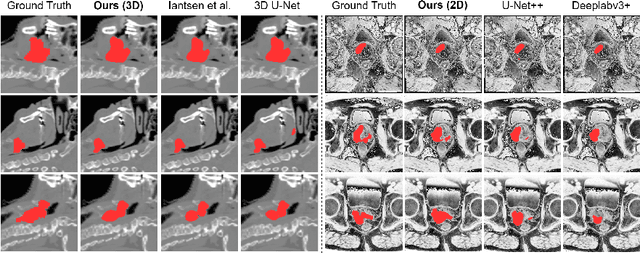
Abstract:Recently, deep learning methods have been widely used for tumor segmentation of multimodal medical images with promising results. However, most existing methods are limited by insufficient representational ability, specific modality number and high computational complexity. In this paper, we propose a hybrid densely connected network for tumor segmentation, named H-DenseFormer, which combines the representational power of the Convolutional Neural Network (CNN) and the Transformer structures. Specifically, H-DenseFormer integrates a Transformer-based Multi-path Parallel Embedding (MPE) module that can take an arbitrary number of modalities as input to extract the fusion features from different modalities. Then, the multimodal fusion features are delivered to different levels of the encoder to enhance multimodal learning representation. Besides, we design a lightweight Densely Connected Transformer (DCT) block to replace the standard Transformer block, thus significantly reducing computational complexity. We conduct extensive experiments on two public multimodal datasets, HECKTOR21 and PI-CAI22. The experimental results show that our proposed method outperforms the existing state-of-the-art methods while having lower computational complexity. The source code is available at https://github.com/shijun18/H-DenseFormer.
Dual-Attention Residual Network for Automatic Diagnosis of COVID-19
May 14, 2021



Abstract:The ongoing global pandemic of Coronavirus Disease 2019 (COVID-19) has posed serious threat to public health and the economy. Rapid and accurate diagnosis of COVID-19 is crucial to prevent the further spread of the disease and reduce its mortality. Chest computed tomography (CT) is an effective tool for the early diagnosis of lung diseases including pneumonia. However, detecting COVID-19 from CT is demanding and prone to human errors as some early-stage patients may have negative findings on images. In this study, we propose a novel residual network to automatically identify COVID-19 from other common pneumonia and normal people using CT images. Specifically, we employ the modified 3D ResNet18 as the backbone network, which is equipped with both channel-wise attention (CA) and depth-wise attention (DA) modules to further improve the diagnostic performance. Experimental results on the large open-source dataset show that our method can differentiate COVID-19 from the other two classes with 94.7% accuracy, 93.73% sensitivity, 98.28% specificity, 95.26% F1-score, and an area under the receiver operating characteristic curve (AUC) of 0.99, outperforming baseline methods. These results demonstrate that the proposed method could potentially assist the clinicians in performing a quick diagnosis to fight COVID-19.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge