Hari Shroff
DeepPD: Joint Phase and Object Estimation from Phase Diversity with Neural Calibration of a Deformable Mirror
Apr 19, 2025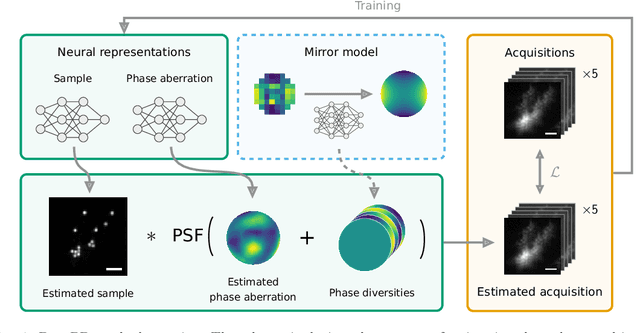


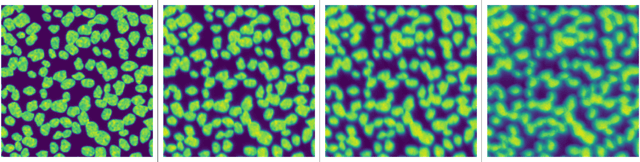
Abstract:Sample-induced aberrations and optical imperfections limit the resolution of fluorescence microscopy. Phase diversity is a powerful technique that leverages complementary phase information in sequentially acquired images with deliberately introduced aberrations--the phase diversities--to enable phase and object reconstruction and restore diffraction-limited resolution. These phase diversities are typically introduced into the optical path via a deformable mirror. Existing phase-diversity-based methods are limited to Zernike modes, require large numbers of diversity images, or depend on accurate mirror calibration--which are all suboptimal. We present DeepPD, a deep learning-based framework that combines neural representations of the object and wavefront with a learned model of the deformable mirror to jointly estimate both object and phase from only five images. DeepPD improves robustness and reconstruction quality over previous approaches, even under severe aberrations. We demonstrate its performance on calibration targets and biological samples, including immunolabeled myosin in fixed PtK2 cells.
Prediction of Cellular Identities from Trajectory and Cell Fate Information
Jan 11, 2024Abstract:Determining cell identities in imaging sequences is an important yet challenging task. The conventional method for cell identification is via cell tracking, which is complex and can be time-consuming. In this study, we propose an innovative approach to cell identification during early C. elegans embryogenesis using machine learning. We employed random forest, MLP, and LSTM models, and tested cell classification accuracy on 3D time-lapse confocal datasets spanning the first 4 hours of embryogenesis. By leveraging a small number of spatial-temporal features of individual cells, including cell trajectory and cell fate information, our models achieve an accuracy of over 90%, even with limited data. We also determine the most important feature contributions and can interpret these features in the context of biological knowledge. Our research demonstrates the success of predicting cell identities in 4D imaging sequences directly from simple spatio-temporal features.
Phase Diverse Phase Retrieval for Microscopy: Comparison of Gaussian and Poisson Approaches
Aug 01, 2023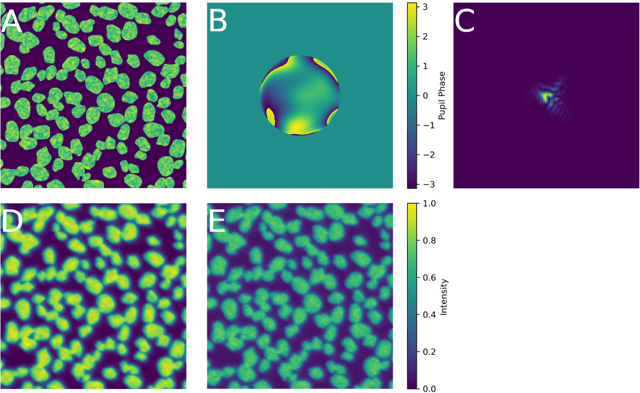


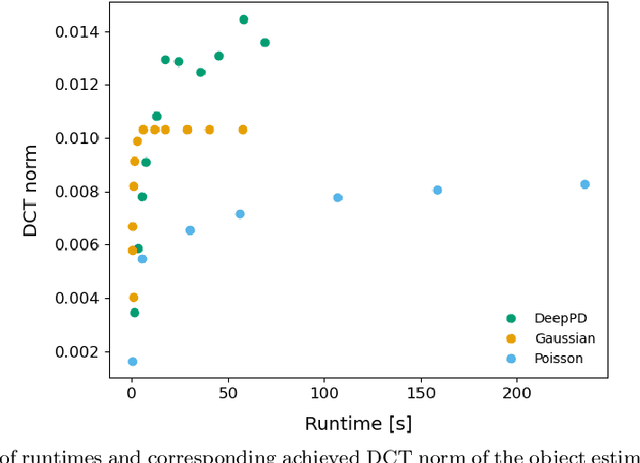
Abstract:Phase diversity is a widefield aberration correction method that uses multiple images to estimate the phase aberration at the pupil plane of an imaging system by solving an optimization problem. This estimated aberration can then be used to deconvolve the aberrated image or to reacquire it with aberration corrections applied to a deformable mirror. The optimization problem for aberration estimation has been formulated for both Gaussian and Poisson noise models but the Poisson model has never been studied in microscopy nor compared with the Gaussian model. Here, the Gaussian- and Poisson-based estimation algorithms are implemented and compared for widefield microscopy in simulation. The Poisson algorithm is found to match or outperform the Gaussian algorithm in a variety of situations, and converges in a similar or decreased amount of time. The Gaussian algorithm does perform better in low-light regimes when image noise is dominated by additive Gaussian noise. The Poisson algorithm is also found to be more robust to the effects of spatially variant aberration and phase noise. Finally, the relative advantages of re-acquisition with aberration correction and deconvolution with aberrated point spread functions are compared.
A Semi-automatic Cell Tracking Process Towards Completing the 4D Atlas of C. elegans Development
Aug 02, 2022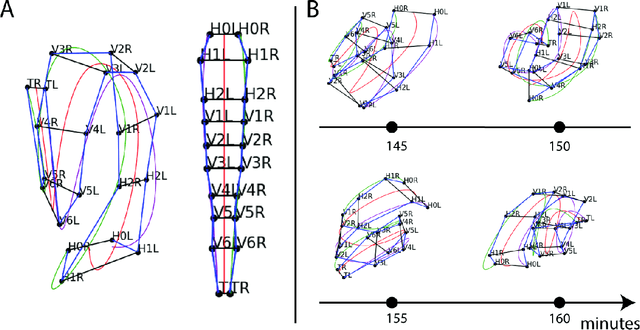
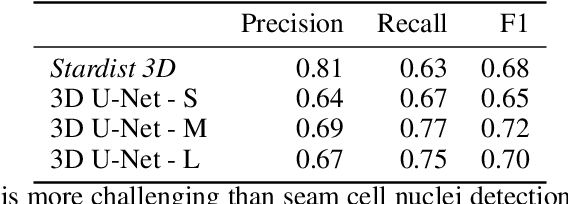
Abstract:The nematode Caenorhabditis elegans (C. elegans) is used as a model organism to better understand developmental biology and neurobiology. C. elegans features an invariant cell lineage, which has been catalogued and observed using fluorescence microscopy images. However, established methods to track cells in late-stage development fail to generalize once sporadic muscular twitching has begun. We build upon methodology which uses skin cells as fiducial markers to carry out cell tracking despite random twitching. In particular, we present a cell nucleus segmentation and tracking procedure which was integrated into a 3D rendering GUI to improve efficiency in tracking cells across late-stage development. Results on images depicting aforementioned muscle cell nuclei across three test embryos suggest the fiducial markers in conjunction with a classic tracking paradigm overcome sporadic twitching.
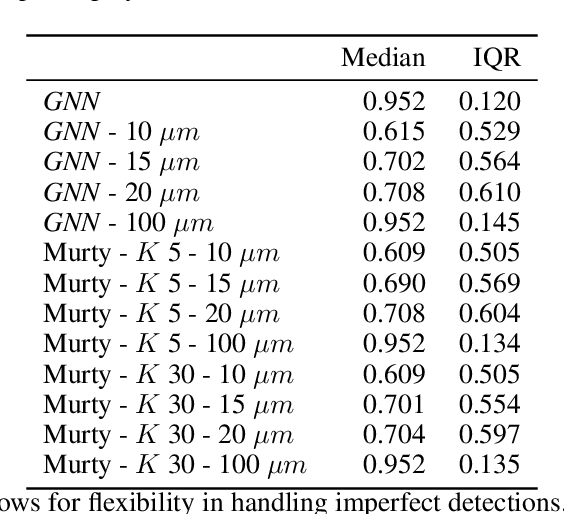
Multiple Hypothesis Hypergraph Tracking for Posture Identification in Embryonic Caenorhabditis elegans
Nov 11, 2021



Abstract:Current methods in multiple object tracking (MOT) rely on independent object trajectories undergoing predictable motion to effectively track large numbers of objects. Adversarial conditions such as volatile object motion and imperfect detections create a challenging tracking landscape in which established methods may yield inadequate results. Multiple hypothesis hypergraph tracking (MHHT) is developed to perform MOT among interdependent objects amid noisy detections. The method extends traditional multiple hypothesis tracking (MHT) via hypergraphs to model correlated object motion, allowing for robust tracking in challenging scenarios. MHHT is applied to perform seam cell tracking during late-stage embryogenesis in embryonic C. elegans.
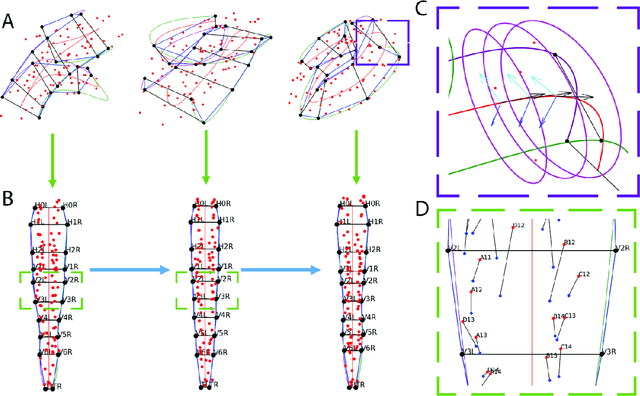
An Exact Hypergraph Matching Algorithm for Nuclear Identification in Embryonic Caenorhabditis elegans
Apr 20, 2021
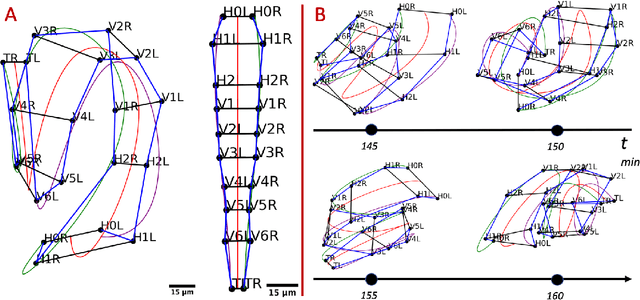
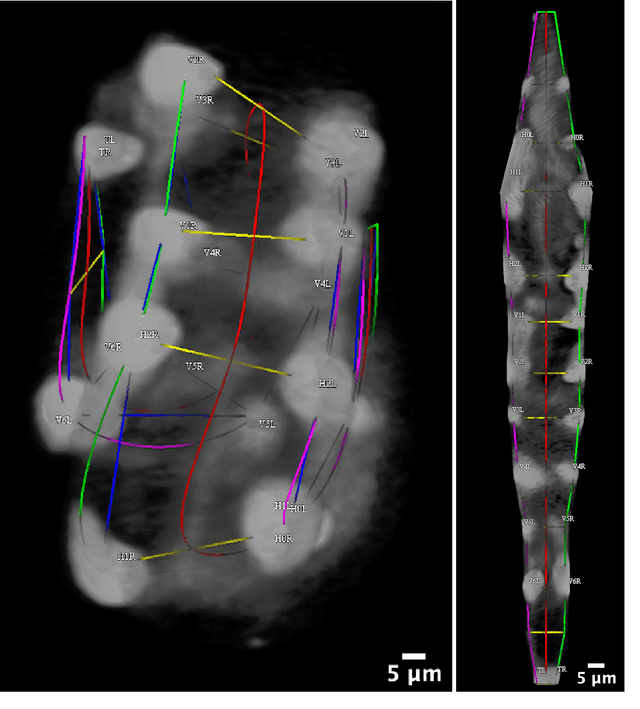

Abstract:Finding an optimal correspondence between point sets is a common task in computer vision. Existing techniques assume relatively simple relationships among points and do not guarantee an optimal match. We introduce an algorithm capable of exactly solving point set matching by modeling the task as hypergraph matching. The algorithm extends the classical branch and bound paradigm to select and aggregate vertices under a proposed decomposition of the multilinear objective function. The methodology is motivated by Caenorhabditis elegans, a model organism used frequently in developmental biology and neurobiology. The embryonic C. elegans contains seam cells that can act as fiducial markers allowing the identification of other nuclei during embryo development. The proposed algorithm identifies seam cells more accurately than established point-set matching methods, while providing a framework to approach other similarly complex point set matching tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge