Ryan Christensen
A Semi-automatic Cell Tracking Process Towards Completing the 4D Atlas of C. elegans Development
Aug 02, 2022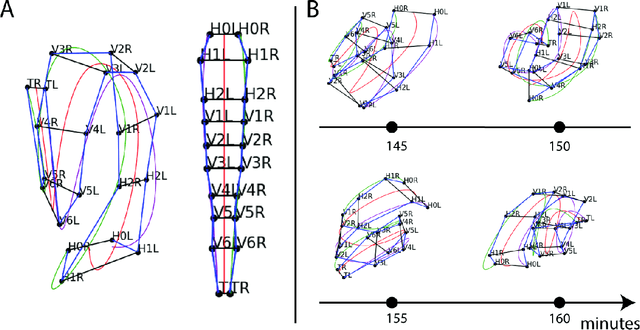
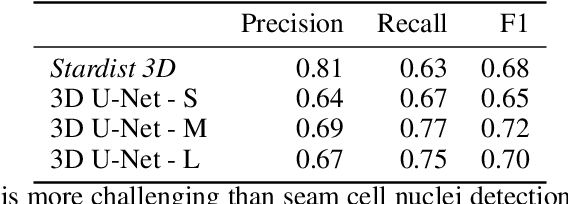
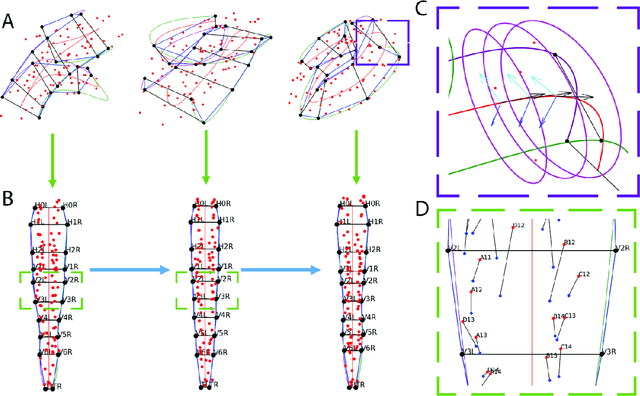
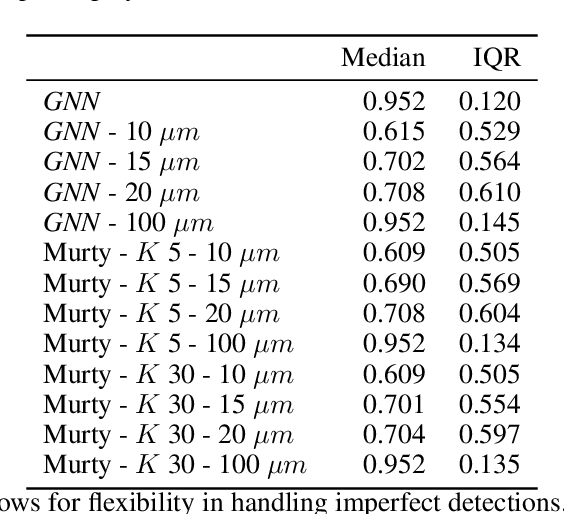
Abstract:The nematode Caenorhabditis elegans (C. elegans) is used as a model organism to better understand developmental biology and neurobiology. C. elegans features an invariant cell lineage, which has been catalogued and observed using fluorescence microscopy images. However, established methods to track cells in late-stage development fail to generalize once sporadic muscular twitching has begun. We build upon methodology which uses skin cells as fiducial markers to carry out cell tracking despite random twitching. In particular, we present a cell nucleus segmentation and tracking procedure which was integrated into a 3D rendering GUI to improve efficiency in tracking cells across late-stage development. Results on images depicting aforementioned muscle cell nuclei across three test embryos suggest the fiducial markers in conjunction with a classic tracking paradigm overcome sporadic twitching.
An Exact Hypergraph Matching Algorithm for Nuclear Identification in Embryonic Caenorhabditis elegans
Apr 20, 2021
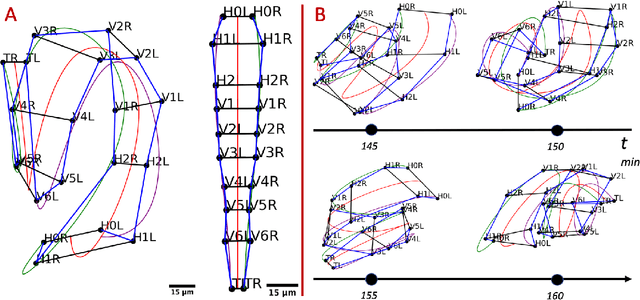
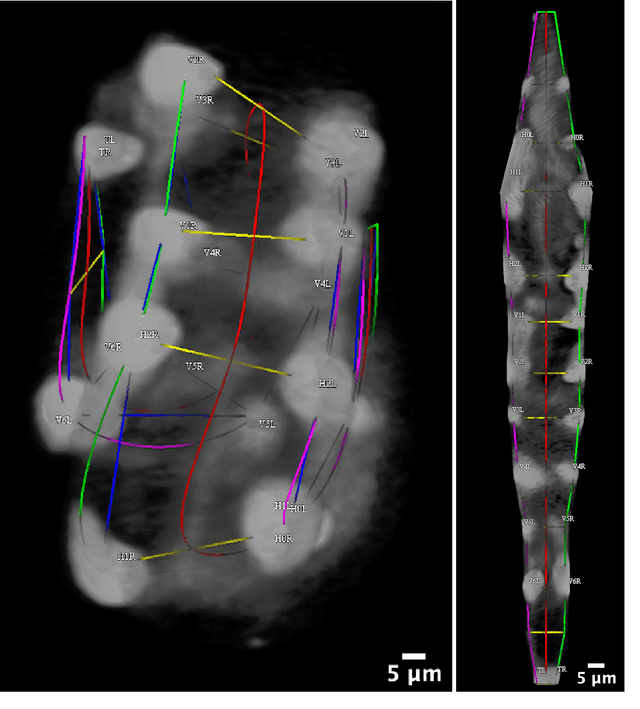

Abstract:Finding an optimal correspondence between point sets is a common task in computer vision. Existing techniques assume relatively simple relationships among points and do not guarantee an optimal match. We introduce an algorithm capable of exactly solving point set matching by modeling the task as hypergraph matching. The algorithm extends the classical branch and bound paradigm to select and aggregate vertices under a proposed decomposition of the multilinear objective function. The methodology is motivated by Caenorhabditis elegans, a model organism used frequently in developmental biology and neurobiology. The embryonic C. elegans contains seam cells that can act as fiducial markers allowing the identification of other nuclei during embryo development. The proposed algorithm identifies seam cells more accurately than established point-set matching methods, while providing a framework to approach other similarly complex point set matching tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge