Haraldur T. Hallgrímsson
Towards Time Series Reasoning with LLMs
Sep 17, 2024Abstract:Multi-modal large language models (MLLMs) have enabled numerous advances in understanding and reasoning in domains like vision, but we have not yet seen this broad success for time-series. Although prior works on time-series MLLMs have shown promising performance in time-series forecasting, very few works show how an LLM could be used for time-series reasoning in natural language. We propose a novel multi-modal time-series LLM approach that learns generalizable information across various domains with powerful zero-shot performance. First, we train a lightweight time-series encoder on top of an LLM to directly extract time-series information. Then, we fine-tune our model with chain-of-thought augmented time-series tasks to encourage the model to generate reasoning paths. We show that our model learns a latent representation that reflects specific time-series features (e.g. slope, frequency), as well as outperforming GPT-4o on a set of zero-shot reasoning tasks on a variety of domains.
Learning Individualized Cardiovascular Responses from Large-scale Wearable Sensors Data
Dec 04, 2018

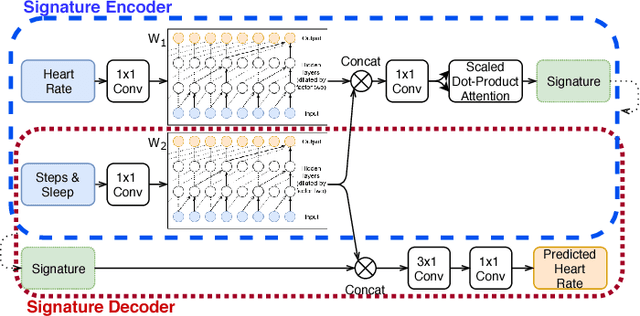
Abstract:We consider the problem of modeling cardiovascular responses to physical activity and sleep changes captured by wearable sensors in free living conditions. We use an attentional convolutional neural network to learn parsimonious signatures of individual cardiovascular response from data recorded at the minute level resolution over several months on a cohort of 80k people. We demonstrate internal validity by showing that signatures generated on an individual's 2017 data generalize to predict minute-level heart rate from physical activity and sleep for the same individual in 2018, outperforming several time-series forecasting baselines. We also show external validity demonstrating that signatures outperform plain resting heart rate (RHR) in predicting variables associated with cardiovascular functions, such as age and Body Mass Index (BMI). We believe that the computed cardiovascular signatures have utility in monitoring cardiovascular health over time, including detecting abnormalities and quantifying recovery from acute events.
Spatial Coherence of Oriented White Matter Microstructure: Applications to White Matter Regions Associated with Genetic Similarity
Feb 14, 2018
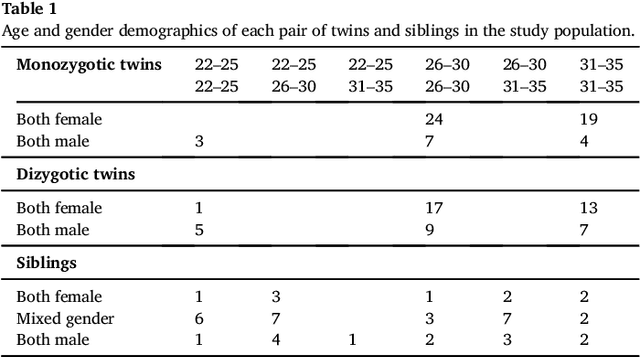


Abstract:We present a method to discover differences between populations with respect to the spatial coherence of their oriented white matter microstructure in arbitrarily shaped white matter regions. This method is applied to diffusion MRI scans of a subset of the Human Connectome Project dataset: 57 pairs of monozygotic and 52 pairs of dizygotic twins. After controlling for morphological similarity between twins, we identify 3.7% of all white matter as being associated with genetic similarity (35.1k voxels, $p < 10^{-4}$, false discovery rate 1.5%), 75% of which spatially clusters into twenty-two contiguous white matter regions. Furthermore, we show that the orientation similarity within these regions generalizes to a subset of 47 pairs of non-twin siblings, and show that these siblings are on average as similar as dizygotic twins. The regions are located in deep white matter including the superior longitudinal fasciculus, the optic radiations, the middle cerebellar peduncle, the corticospinal tract, and within the anterior temporal lobe, as well as the cerebellum, brain stem, and amygdalae. These results extend previous work using undirected fractional anisotrophy for measuring putative heritable influences in white matter. Our multidirectional extension better accounts for crossing fiber connections within voxels. This bottom up approach has at its basis a novel measurement of coherence within neighboring voxel dyads between subjects, and avoids some of the fundamental ambiguities encountered with tractographic approaches to white matter analysis that estimate global connectivity.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge