Spatial Coherence of Oriented White Matter Microstructure: Applications to White Matter Regions Associated with Genetic Similarity
Paper and Code
Feb 14, 2018
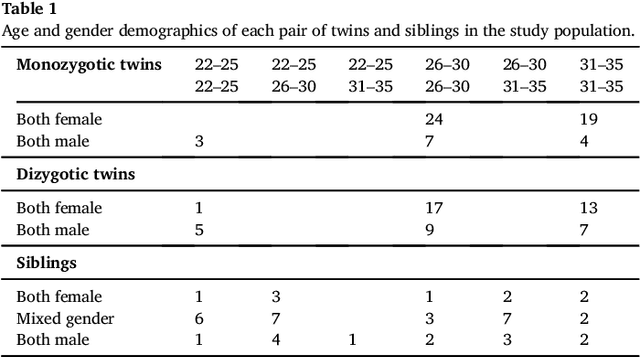


We present a method to discover differences between populations with respect to the spatial coherence of their oriented white matter microstructure in arbitrarily shaped white matter regions. This method is applied to diffusion MRI scans of a subset of the Human Connectome Project dataset: 57 pairs of monozygotic and 52 pairs of dizygotic twins. After controlling for morphological similarity between twins, we identify 3.7% of all white matter as being associated with genetic similarity (35.1k voxels, $p < 10^{-4}$, false discovery rate 1.5%), 75% of which spatially clusters into twenty-two contiguous white matter regions. Furthermore, we show that the orientation similarity within these regions generalizes to a subset of 47 pairs of non-twin siblings, and show that these siblings are on average as similar as dizygotic twins. The regions are located in deep white matter including the superior longitudinal fasciculus, the optic radiations, the middle cerebellar peduncle, the corticospinal tract, and within the anterior temporal lobe, as well as the cerebellum, brain stem, and amygdalae. These results extend previous work using undirected fractional anisotrophy for measuring putative heritable influences in white matter. Our multidirectional extension better accounts for crossing fiber connections within voxels. This bottom up approach has at its basis a novel measurement of coherence within neighboring voxel dyads between subjects, and avoids some of the fundamental ambiguities encountered with tractographic approaches to white matter analysis that estimate global connectivity.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge