Gustavo Malkomes
Group SELFIES: A Robust Fragment-Based Molecular String Representation
Nov 23, 2022


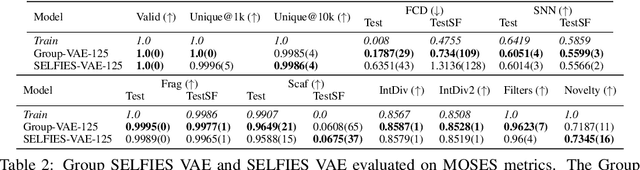
Abstract:We introduce Group SELFIES, a molecular string representation that leverages group tokens to represent functional groups or entire substructures while maintaining chemical robustness guarantees. Molecular string representations, such as SMILES and SELFIES, serve as the basis for molecular generation and optimization in chemical language models, deep generative models, and evolutionary methods. While SMILES and SELFIES leverage atomic representations, Group SELFIES builds on top of the chemical robustness guarantees of SELFIES by enabling group tokens, thereby creating additional flexibility to the representation. Moreover, the group tokens in Group SELFIES can take advantage of inductive biases of molecular fragments that capture meaningful chemical motifs. The advantages of capturing chemical motifs and flexibility are demonstrated in our experiments, which show that Group SELFIES improves distribution learning of common molecular datasets. Further experiments also show that random sampling of Group SELFIES strings improves the quality of generated molecules compared to regular SELFIES strings. Our open-source implementation of Group SELFIES is available online, which we hope will aid future research in molecular generation and optimization.
Accelerating Psychometric Screening Tests With Bayesian Active Differential Selection
Feb 04, 2020



Abstract:Classical methods for psychometric function estimation either require excessive measurements or produce only a low-resolution approximation of the target psychometric function. In this paper, we propose a novel solution for rapid screening for a change in the psychometric function estimation of a given patient. We use Bayesian active model selection to perform an automated pure-tone audiogram test with the goal of quickly finding if the current audiogram will be different from a previous audiogram. We validate our approach using audiometric data from the National Institute for Occupational Safety and Health NIOSH. Initial results show that with a few tones we can detect if the patient's audiometric function has changed between the two test sessions with high confidence.
Efficient nonmyopic active search with applications in drug and materials discovery
Nov 23, 2018



Abstract:Active search is a learning paradigm for actively identifying as many members of a given class as possible. A critical target scenario is high-throughput screening for scientific discovery, such as drug or materials discovery. In this paper, we approach this problem in Bayesian decision framework. We first derive the Bayesian optimal policy under a natural utility, and establish a theoretical hardness of active search, proving that the optimal policy can not be approximated for any constant ratio. We also study the batch setting for the first time, where a batch of $b>1$ points can be queried at each iteration. We give an asymptotic lower bound, linear in batch size, on the adaptivity gap: how much we could lose if we query $b$ points at a time for $t$ iterations, instead of one point at a time for $bt$ iterations. We then introduce a novel approach to nonmyopic approximations of the optimal policy that admits efficient computation. Our proposed policy can automatically trade off exploration and exploitation, without relying on any tuning parameters. We also generalize our policy to batch setting, and propose two approaches to tackle the combinatorial search challenge. We evaluate our proposed policies on a large database of drug discovery and materials science. Results demonstrate the superior performance of our proposed policy in both sequential and batch setting; the nonmyopic behavior is also illustrated in various aspects.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge