Guoan Zhang
AACR-Bench: Evaluating Automatic Code Review with Holistic Repository-Level Context
Jan 27, 2026Abstract:High-quality evaluation benchmarks are pivotal for deploying Large Language Models (LLMs) in Automated Code Review (ACR). However, existing benchmarks suffer from two critical limitations: first, the lack of multi-language support in repository-level contexts, which restricts the generalizability of evaluation results; second, the reliance on noisy, incomplete ground truth derived from raw Pull Request (PR) comments, which constrains the scope of issue detection. To address these challenges, we introduce AACR-Bench a comprehensive benchmark that provides full cross-file context across multiple programming languages. Unlike traditional datasets, AACR-Bench employs an "AI-assisted, Expert-verified" annotation pipeline to uncover latent defects often overlooked in original PRs, resulting in a 285\% increase in defect coverage. Extensive evaluations of mainstream LLMs on AACR-Bench reveal that previous assessments may have either misjudged or only partially captured model capabilities due to data limitations. Our work establishes a more rigorous standard for ACR evaluation and offers new insights on LLM based ACR, i.e., the granularity/level of context and the choice of retrieval methods significantly impact ACR performance, and this influence varies depending on the LLM, programming language, and the LLM usage paradigm e.g., whether an Agent architecture is employed. The code, data, and other artifacts of our evaluation set are available at https://github.com/alibaba/aacr-bench .
M2rc-Eval: Massively Multilingual Repository-level Code Completion Evaluation
Oct 28, 2024



Abstract:Repository-level code completion has drawn great attention in software engineering, and several benchmark datasets have been introduced. However, existing repository-level code completion benchmarks usually focus on a limited number of languages (<5), which cannot evaluate the general code intelligence abilities across different languages for existing code Large Language Models (LLMs). Besides, the existing benchmarks usually report overall average scores of different languages, where the fine-grained abilities in different completion scenarios are ignored. Therefore, to facilitate the research of code LLMs in multilingual scenarios, we propose a massively multilingual repository-level code completion benchmark covering 18 programming languages (called M2RC-EVAL), and two types of fine-grained annotations (i.e., bucket-level and semantic-level) on different completion scenarios are provided, where we obtain these annotations based on the parsed abstract syntax tree. Moreover, we also curate a massively multilingual instruction corpora M2RC- INSTRUCT dataset to improve the repository-level code completion abilities of existing code LLMs. Comprehensive experimental results demonstrate the effectiveness of our M2RC-EVAL and M2RC-INSTRUCT.
BioCopy: A Plug-And-Play Span Copy Mechanism in Seq2Seq Models
Sep 26, 2021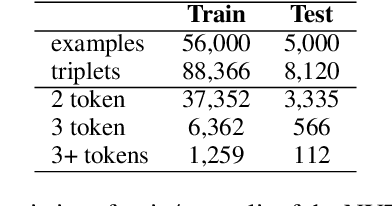
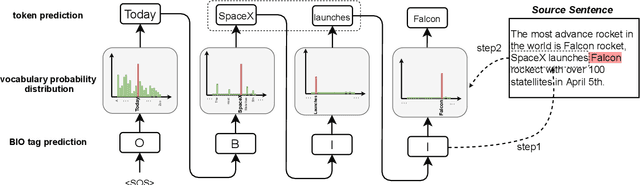
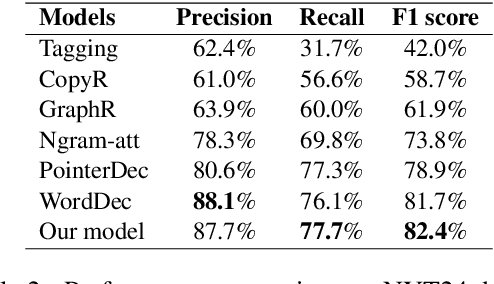
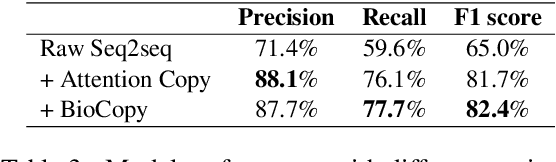
Abstract:Copy mechanisms explicitly obtain unchanged tokens from the source (input) sequence to generate the target (output) sequence under the neural seq2seq framework. However, most of the existing copy mechanisms only consider single word copying from the source sentences, which results in losing essential tokens while copying long spans. In this work, we propose a plug-and-play architecture, namely BioCopy, to alleviate the problem aforementioned. Specifically, in the training stage, we construct a BIO tag for each token and train the original model with BIO tags jointly. In the inference stage, the model will firstly predict the BIO tag at each time step, then conduct different mask strategies based on the predicted BIO label to diminish the scope of the probability distributions over the vocabulary list. Experimental results on two separate generative tasks show that they all outperform the baseline models by adding our BioCopy to the original model structure.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge