Farida Cheriet
Mesh based segmentation for automated margin line generation on incisors receiving crown treatment
Jul 30, 2025



Abstract:Dental crowns are essential dental treatments for restoring damaged or missing teeth of patients. Recent design approaches of dental crowns are carried out using commercial dental design software. Once a scan of a preparation is uploaded to the software, a dental technician needs to manually define a precise margin line on the preparation surface, which constitutes a non-repeatable and inconsistent procedure. This work proposes a new framework to determine margin lines automatically and accurately using deep learning. A dataset of incisor teeth was provided by a collaborating dental laboratory to train a deep learning segmentation model. A mesh-based neural network was modified by changing its input channels and used to segment the prepared tooth into two regions such that the margin line is contained within the boundary faces separating the two regions. Next, k-fold cross-validation was used to train 5 models, and a voting classifier technique was used to combine their results to enhance the segmentation. After that, boundary smoothing and optimization using the graph cut method were applied to refine the segmentation results. Then, boundary faces separating the two regions were selected to represent the margin line faces. A spline was approximated to best fit the centers of the boundary faces to predict the margin line. Our results show that an ensemble model combined with maximum probability predicted the highest number of successful test cases (7 out of 13) based on a maximum distance threshold of 200 m (representing human error) between the predicted and ground truth point clouds. It was also demonstrated that the better the quality of the preparation, the smaller the divergence between the predicted and ground truth margin lines (Spearman's rank correlation coefficient of -0.683). We provide the train and test datasets for the community.
From Mesh Completion to AI Designed Crown
Jan 09, 2025Abstract:Designing a dental crown is a time-consuming and labor intensive process. Our goal is to simplify crown design and minimize the tediousness of making manual adjustments while still ensuring the highest level of accuracy and consistency. To this end, we present a new end- to-end deep learning approach, coined Dental Mesh Completion (DMC), to generate a crown mesh conditioned on a point cloud context. The dental context includes the tooth prepared to receive a crown and its surroundings, namely the two adjacent teeth and the three closest teeth in the opposing jaw. We formulate crown generation in terms of completing this point cloud context. A feature extractor first converts the input point cloud into a set of feature vectors that represent local regions in the point cloud. The set of feature vectors is then fed into a transformer to predict a new set of feature vectors for the missing region (crown). Subsequently, a point reconstruction head, followed by a multi-layer perceptron, is used to predict a dense set of points with normals. Finally, a differentiable point-to-mesh layer serves to reconstruct the crown surface mesh. We compare our DMC method to a graph-based convolutional neural network which learns to deform a crown mesh from a generic crown shape to the target geometry. Extensive experiments on our dataset demonstrate the effectiveness of our method, which attains an average of 0.062 Chamfer Distance.The code is available at:https://github.com/Golriz-code/DMC.gi
Multi-style conversion for semantic segmentation of lesions in fundus images by adversarial attacks
Oct 17, 2024Abstract:The diagnosis of diabetic retinopathy, which relies on fundus images, faces challenges in achieving transparency and interpretability when using a global classification approach. However, segmentation-based databases are significantly more expensive to acquire and combining them is often problematic. This paper introduces a novel method, termed adversarial style conversion, to address the lack of standardization in annotation styles across diverse databases. By training a single architecture on combined databases, the model spontaneously modifies its segmentation style depending on the input, demonstrating the ability to convert among different labeling styles. The proposed methodology adds a linear probe to detect dataset origin based on encoder features and employs adversarial attacks to condition the model's segmentation style. Results indicate significant qualitative and quantitative through dataset combination, offering avenues for improved model generalization, uncertainty estimation and continuous interpolation between annotation styles. Our approach enables training a segmentation model with diverse databases while controlling and leveraging annotation styles for improved retinopathy diagnosis.
Cross-Dataset Generalization For Retinal Lesions Segmentation
May 14, 2024



Abstract:Identifying lesions in fundus images is an important milestone toward an automated and interpretable diagnosis of retinal diseases. To support research in this direction, multiple datasets have been released, proposing groundtruth maps for different lesions. However, important discrepancies exist between the annotations and raise the question of generalization across datasets. This study characterizes several known datasets and compares different techniques that have been proposed to enhance the generalisation performance of a model, such as stochastic weight averaging, model soups and ensembles. Our results provide insights into how to combine coarsely labelled data with a finely-grained dataset in order to improve the lesions segmentation.
MAPLES-DR: MESSIDOR Anatomical and Pathological Labels for Explainable Screening of Diabetic Retinopathy
Jan 19, 2024Abstract:Reliable automatic diagnosis of Diabetic Retinopathy (DR) and Macular Edema (ME) is an invaluable asset in improving the rate of monitored patients among at-risk populations and in enabling earlier treatments before the pathology progresses and threatens vision. However, the explainability of screening models is still an open question, and specifically designed datasets are required to support the research. We present MAPLES-DR (MESSIDOR Anatomical and Pathological Labels for Explainable Screening of Diabetic Retinopathy), which contains, for 198 images of the MESSIDOR public fundus dataset, new diagnoses for DR and ME as well as new pixel-wise segmentation maps for 10 anatomical and pathological biomarkers related to DR. This paper documents the design choices and the annotation procedure that produced MAPLES-DR, discusses the interobserver variability and the overall quality of the annotations, and provides guidelines on using the dataset in a machine learning context.
Multi-domain learning CNN model for microscopy image classification
Apr 20, 2023Abstract:For any type of microscopy image, getting a deep learning model to work well requires considerable effort to select a suitable architecture and time to train it. As there is a wide range of microscopes and experimental setups, designing a single model that can apply to multiple imaging domains, instead of having multiple per-domain models, becomes more essential. This task is challenging and somehow overlooked in the literature. In this paper, we present a multi-domain learning architecture for the classification of microscopy images that differ significantly in types and contents. Unlike previous methods that are computationally intensive, we have developed a compact model, called Mobincep, by combining the simple but effective techniques of depth-wise separable convolution and the inception module. We also introduce a new optimization technique to regulate the latent feature space during training to improve the network's performance. We evaluated our model on three different public datasets and compared its performance in single-domain and multiple-domain learning modes. The proposed classifier surpasses state-of-the-art results and is robust for limited labeled data. Moreover, it helps to eliminate the burden of designing a new network when switching to new experiments.
Improving the quality of dental crown using a Transformer-based method
Mar 04, 2023Abstract:Designing a synthetic crown is a time-consuming, inconsistent, and labor-intensive process. In this work, we present a fully automatic method that not only learns human design dental crowns, but also improves the consistency, functionality, and esthetic of the crowns. Following success in point cloud completion using the transformer-based network, we tackle the problem of the crown generation as a point-cloud completion around a prepared tooth. To this end, we use a geometry-aware transformer to generate dental crowns. Our main contribution is to add a margin line information to the network, as the accuracy of generating a precise margin line directly,determines whether the designed crown and prepared tooth are closely matched to allowappropriateadhesion.Using our ground truth crown, we can extract the margin line as a spline and sample the spline into 1000 points. We feed the obtained margin line along with two neighbor teeth of the prepared tooth and three closest teeth in the opposing jaw. We also add the margin line points to our ground truth crown to increase the resolution at the margin line. Our experimental results show an improvement in the quality of the designed crown when considering the actual context composed of the prepared tooth along with the margin line compared with a crown generated in an empty space as was done by other studies in the literature.
Semi-supervised segmentation of tooth from 3D Scanned Dental Arches
Aug 10, 2022Abstract:Teeth segmentation is an important topic in dental restorations that is essential for crown generation, diagnosis, and treatment planning. In the dental field, the variability of input data is high and there are no publicly available 3D dental arch datasets. Although there has been improvement in the field provided by recent deep learning architectures on 3D data, there still exists some problems such as properly identifying missing teeth in an arch. We propose to use spectral clustering as a self-supervisory signal to joint-train neural networks for segmentation of 3D arches. Our approach is motivated by the observation that K-means clustering provides cues to capture margin lines related to human perception. The main idea is to automatically generate training data by decomposing unlabeled 3D arches into segments relying solely on geometric information. The network is then trained using a joint loss that combines a supervised loss of annotated input and a self-supervised loss of non-labeled input. Our collected data has a variety of arches including arches with missing teeth. Our experimental results show improvement over the fully supervised state-of-the-art MeshSegNet when using semi-supervised learning. Finally, we contribute code and a dataset.
Adaptable Deformable Convolutions for Semantic Segmentation of Fisheye Images in Autonomous Driving Systems
Feb 19, 2021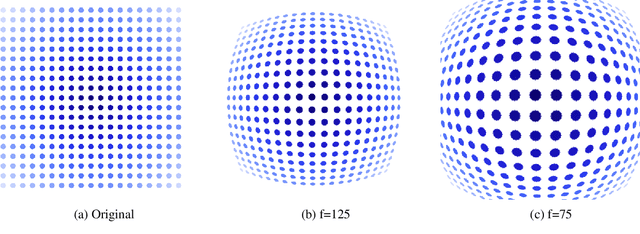
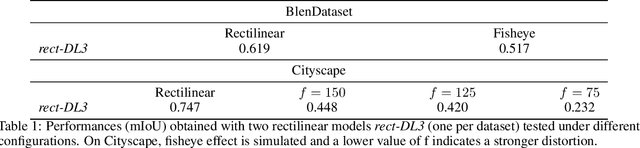


Abstract:Advanced Driver-Assistance Systems rely heavily on perception tasks such as semantic segmentation where images are captured from large field of view (FoV) cameras. State-of-the-art works have made considerable progress toward applying Convolutional Neural Network (CNN) to standard (rectilinear) images. However, the large FoV cameras used in autonomous vehicles produce fisheye images characterized by strong geometric distortion. This work demonstrates that a CNN trained on standard images can be readily adapted to fisheye images, which is crucial in real-world applications where time-consuming real-time data transformation must be avoided. Our adaptation protocol mainly relies on modifying the support of the convolutions by using their deformable equivalents on top of pre-existing layers. We prove that tuning an optimal support only requires a limited amount of labeled fisheye images, as a small number of training samples is sufficient to significantly improve an existing model's performance on wide-angle images. Furthermore, we show that finetuning the weights of the network is not necessary to achieve high performance once the deformable components are learned. Finally, we provide an in-depth analysis of the effect of the deformable convolutions, bringing elements of discussion on the behavior of CNN models.
Three-dimensional Segmentation of the Scoliotic Spine from MRI using Unsupervised Volume-based MR-CT Synthesis
Nov 25, 2020



Abstract:Vertebral bone segmentation from magnetic resonance (MR) images is a challenging task. Due to the inherent nature of the modality to emphasize soft tissues of the body, common thresholding algorithms are ineffective in detecting bones in MR images. On the other hand, it is relatively easier to segment bones from CT images because of the high contrast between bones and the surrounding regions. For this reason, we perform a cross-modality synthesis between MR and CT domains for simple thresholding-based segmentation of the vertebral bones. However, this implicitly assumes the availability of paired MR-CT data, which is rare, especially in the case of scoliotic patients. In this paper, we present a completely unsupervised, fully three-dimensional (3D) cross-modality synthesis method for segmenting scoliotic spines. A 3D CycleGAN model is trained for an unpaired volume-to-volume translation across MR and CT domains. Then, the Otsu thresholding algorithm is applied to the synthesized CT volumes for easy segmentation of the vertebral bones. The resulting segmentation is used to reconstruct a 3D model of the spine. We validate our method on 28 scoliotic vertebrae in 3 patients by computing the point-to-surface mean distance between the landmark points for each vertebra obtained from pre-operative X-rays and the surface of the segmented vertebra. Our study results in a mean error of 3.41 $\pm$ 1.06 mm. Based on qualitative and quantitative results, we conclude that our method is able to obtain a good segmentation and 3D reconstruction of scoliotic spines, all after training from unpaired data in an unsupervised manner.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge