Evan Schwab
Fully Automated CTC Detection, Segmentation and Classification for Multi-Channel IF Imaging
Oct 03, 2024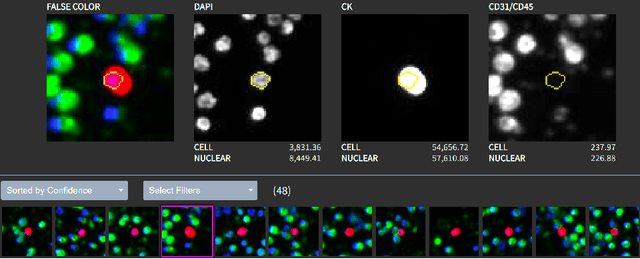
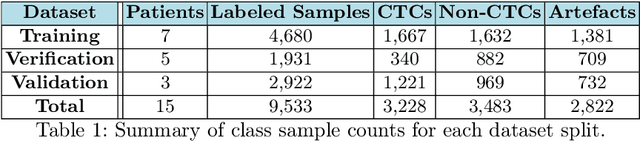
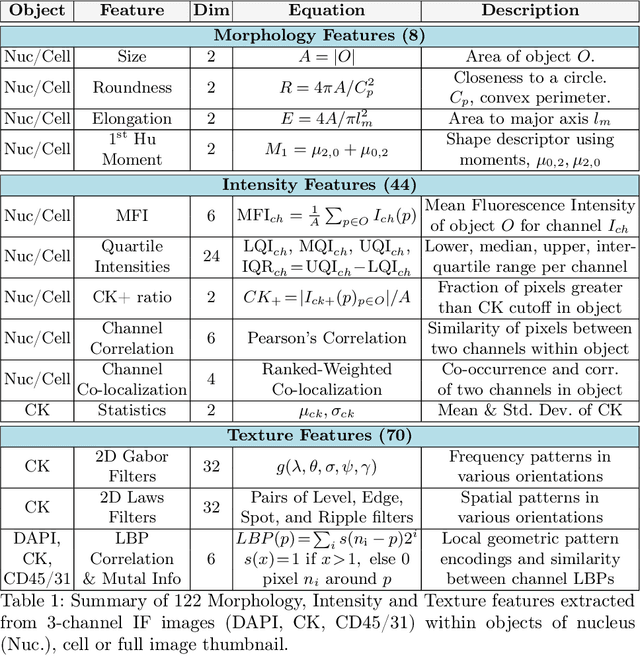
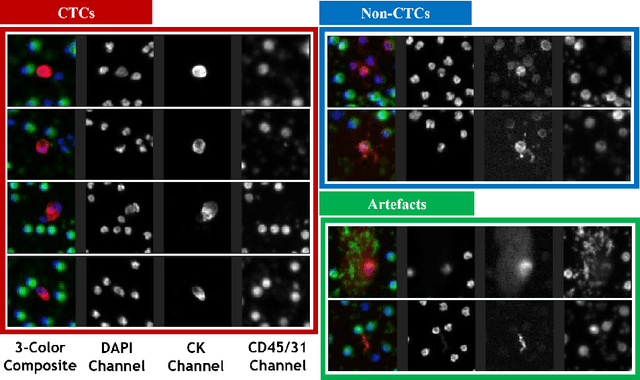
Abstract:Liquid biopsies (eg., blood draws) offer a less invasive and non-localized alternative to tissue biopsies for monitoring the progression of metastatic breast cancer (mBCa). Immunofluoresence (IF) microscopy is a tool to image and analyze millions of blood cells in a patient sample. By detecting and genetically sequencing circulating tumor cells (CTCs) in the blood, personalized treatment plans are achievable for various cancer subtypes. However, CTCs are rare (about 1 in 2M), making manual CTC detection very difficult. In addition, clinicians rely on quantitative cellular biomarkers to manually classify CTCs. This requires prior tasks of cell detection, segmentation and feature extraction. To assist clinicians, we have developed a fully automated machine learning-based production-level pipeline to efficiently detect, segment and classify CTCs in multi-channel IF images. We achieve over 99% sensitivity and 97% specificity on 9,533 cells from 15 mBCa patients. Our pipeline has been successfully deployed on real mBCa patients, reducing a patient average of 14M detected cells to only 335 CTC candidates for manual review.
Automatic Estimation of Ulcerative Colitis Severity from Endoscopy Videos using Ordinal Multi-Instance Learning
Sep 29, 2021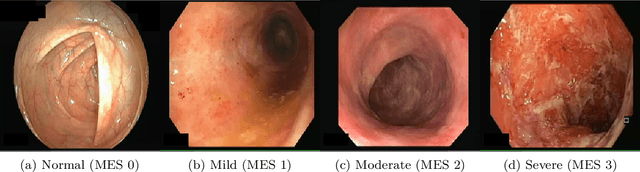
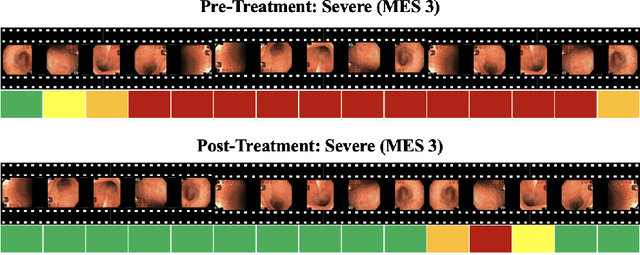
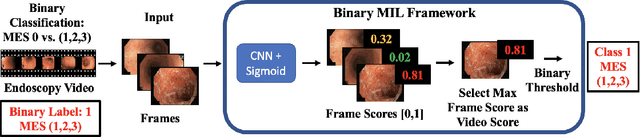
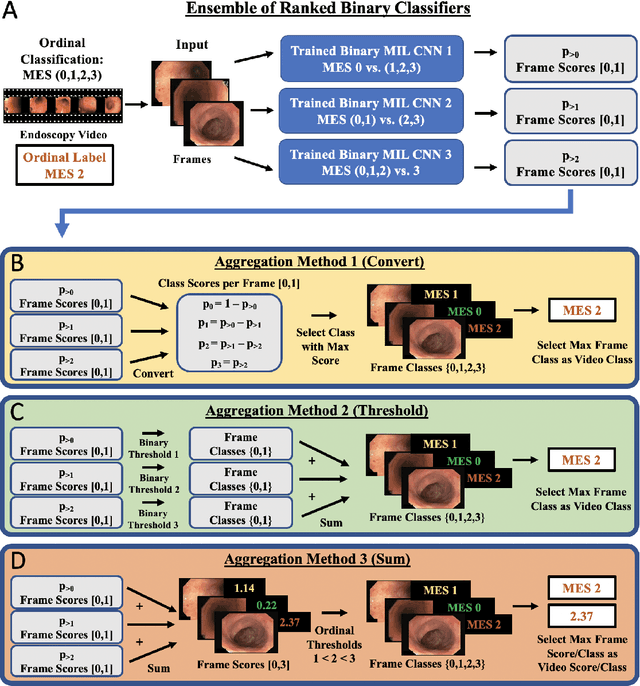
Abstract:Ulcerative colitis (UC) is a chronic inflammatory bowel disease characterized by relapsing inflammation of the large intestine. The severity of UC is often represented by the Mayo Endoscopic Subscore (MES) which quantifies mucosal disease activity from endoscopy videos. In clinical trials, an endoscopy video is assigned an MES based upon the most severe disease activity observed in the video. For this reason, severe inflammation spread throughout the colon will receive the same MES as an otherwise healthy colon with severe inflammation restricted to a small, localized segment. Therefore, the extent of disease activity throughout the large intestine, and overall response to treatment, may not be completely captured by the MES. In this work, we aim to automatically estimate UC severity for each frame in an endoscopy video to provide a higher resolution assessment of disease activity throughout the colon. Because annotating severity at the frame-level is expensive, labor-intensive, and highly subjective, we propose a novel weakly supervised, ordinal classification method to estimate frame severity from video MES labels alone. Using clinical trial data, we first achieved 0.92 and 0.90 AUC for predicting mucosal healing and remission of UC, respectively. Then, for severity estimation, we demonstrate that our models achieve substantial Cohen's Kappa agreement with ground truth MES labels, comparable to the inter-rater agreement of expert clinicians. These findings indicate that our framework could serve as a foundation for novel clinical endpoints, based on a more localized scoring system, to better evaluate UC drug efficacy in clinical trials.
Localization of Critical Findings in Chest X-Ray without Local Annotations Using Multi-Instance Learning
Jan 23, 2020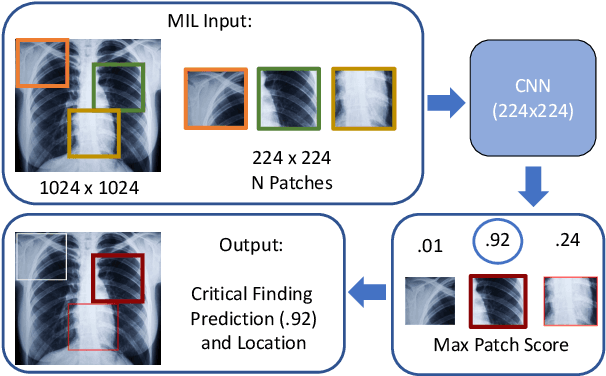

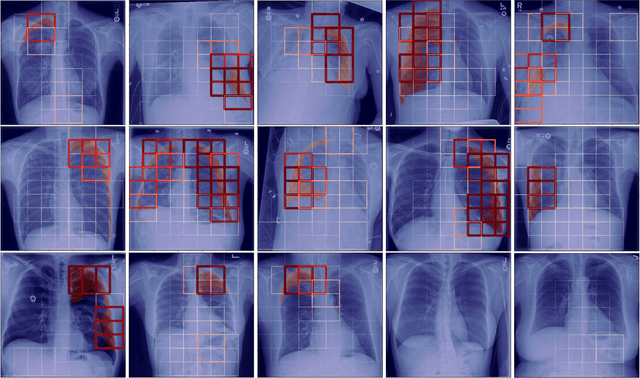
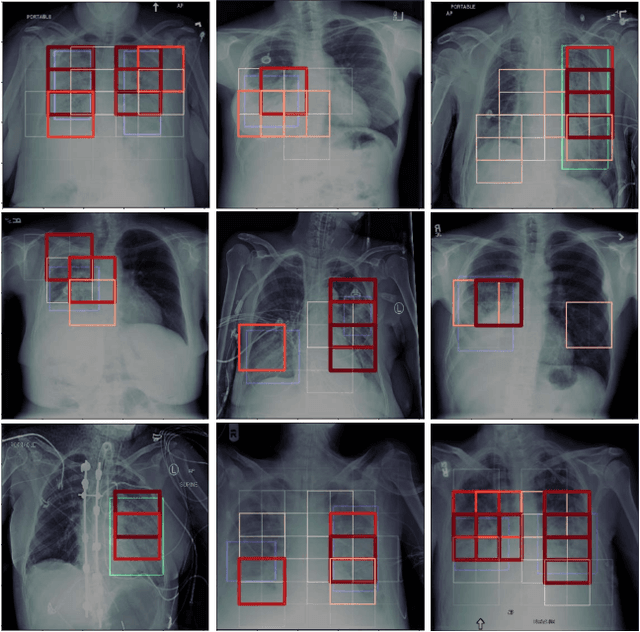
Abstract:The automatic detection of critical findings in chest X-rays (CXR), such as pneumothorax, is important for assisting radiologists in their clinical workflow like triaging time-sensitive cases and screening for incidental findings. While deep learning (DL) models has become a promising predictive technology with near-human accuracy, they commonly suffer from a lack of explainability, which is an important aspect for clinical deployment of DL models in the highly regulated healthcare industry. For example, localizing critical findings in an image is useful for explaining the predictions of DL classification algorithms. While there have been a host of joint classification and localization methods for computer vision, the state-of-the-art DL models require locally annotated training data in the form of pixel level labels or bounding box coordinates. In the medical domain, this requires an expensive amount of manual annotation by medical experts for each critical finding. This requirement becomes a major barrier for training models that can rapidly scale to various findings. In this work, we address these shortcomings with an interpretable DL algorithm based on multi-instance learning that jointly classifies and localizes critical findings in CXR without the need for local annotations. We show competitive classification results on three different critical findings (pneumothorax, pneumonia, and pulmonary edema) from three different CXR datasets.
Deep Learning for Pneumothorax Detection and Localization in Chest Radiographs
Jul 16, 2019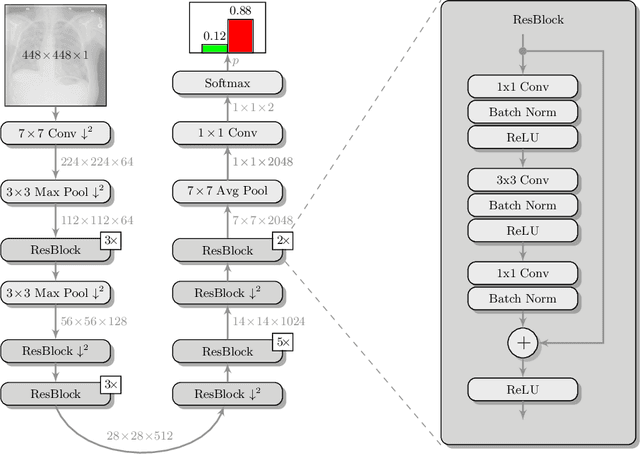
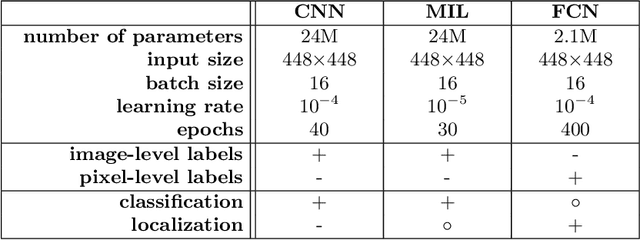
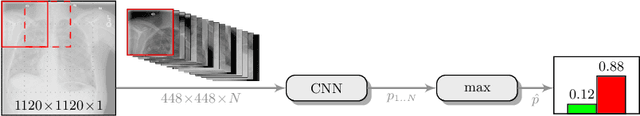
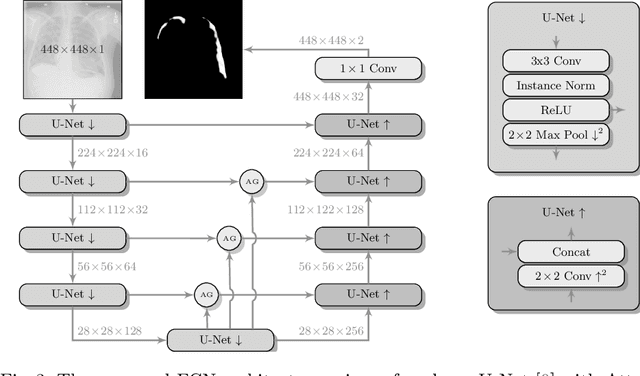
Abstract:Pneumothorax is a critical condition that requires timely communication and immediate action. In order to prevent significant morbidity or patient death, early detection is crucial. For the task of pneumothorax detection, we study the characteristics of three different deep learning techniques: (i) convolutional neural networks, (ii) multiple-instance learning, and (iii) fully convolutional networks. We perform a five-fold cross-validation on a dataset consisting of 1003 chest X-ray images. ROC analysis yields AUCs of 0.96, 0.93, and 0.92 for the three methods, respectively. We review the classification and localization performance of these approaches as well as an ensemble of the three aforementioned techniques.
Separable Dictionary Learning with Global Optimality and Applications to Diffusion MRI
Jul 15, 2018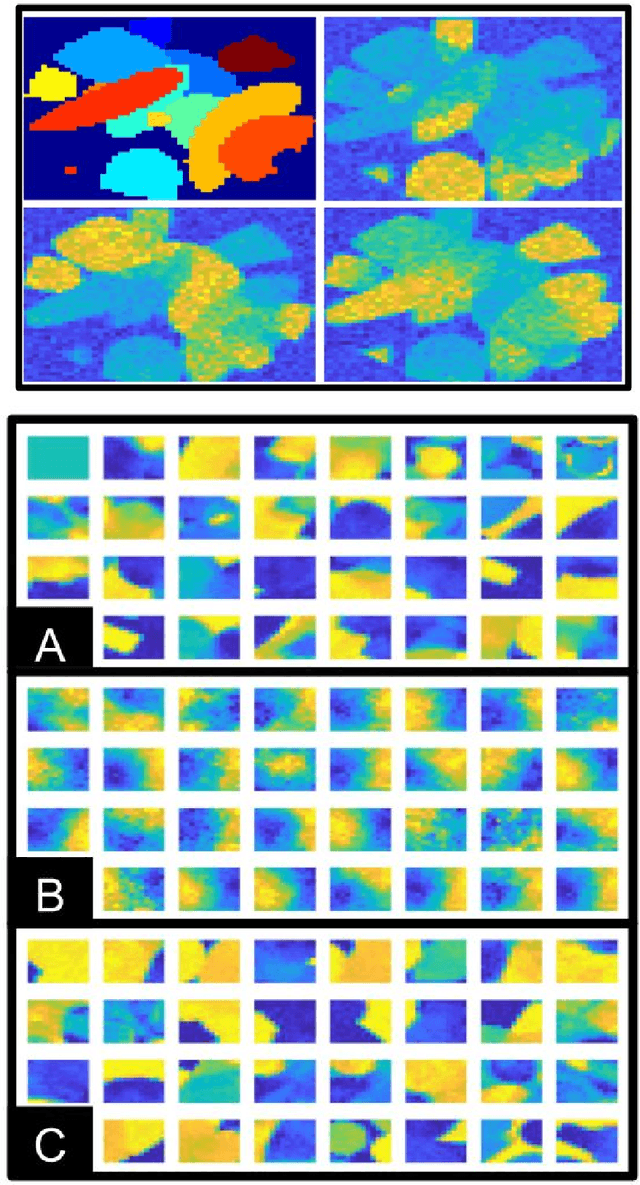

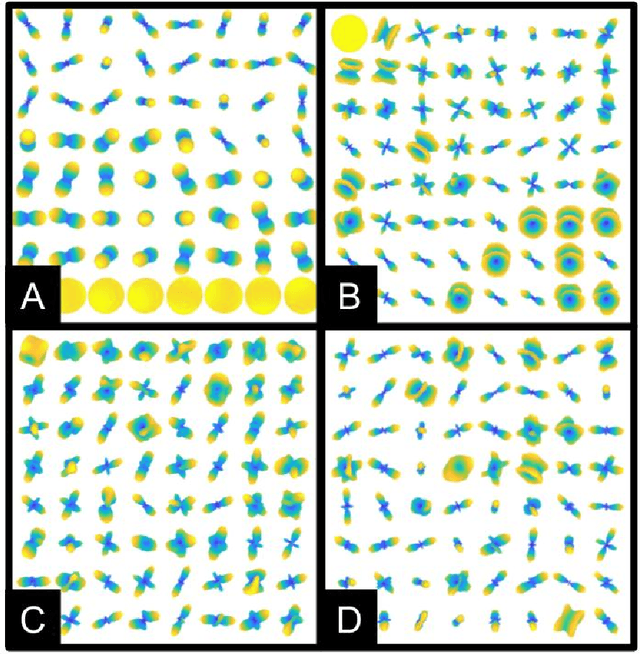

Abstract:Dictionary learning is a popular class of methods for modeling complex data by learning sparse representations directly from the data. For some large-scale applications, exploiting a known structure of the signal is often essential for reducing the complexity of algorithms and representations. One such method is tensor factorization by which a large multi-dimensional dataset can be explicitly factored or separated along each dimension of the data in order to break the representation up into smaller components. Learning dictionaries for tensor structured data is called tensor or separable dictionary learning. While there have been many recent works on separable dictionary learning, typical formulations involve solving a non-convex optimization problem and guaranteeing global optimality remains a challenge. In this work, we propose a framework that uses recent developments in matrix/tensor factorization to provide theoretical and numerical guarantees of the global optimality for the separable dictionary learning problem. We will demonstrate our algorithm on diffusion magnetic resonance imaging (dMRI) data, a medical imaging modality which measures water diffusion along multiple angular directions in every voxel of an MRI volume. For this application, state-of-the-art methods learn dictionaries for the angular domain of the signals without consideration for the spatial domain. In this work, we apply the proposed separable dictionary learning method to learn spatial and angular dMRI dictionaries jointly and show results on denoising phantom and real dMRI brain data.
Joint Spatial-Angular Sparse Coding for dMRI with Separable Dictionaries
May 29, 2018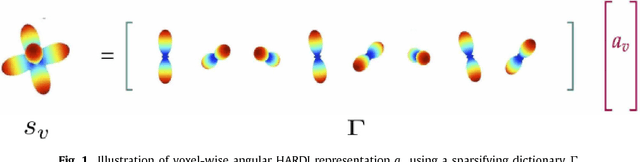
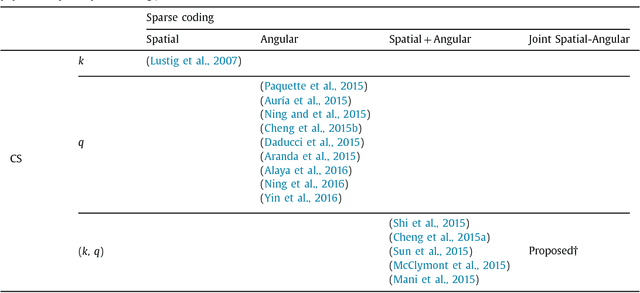

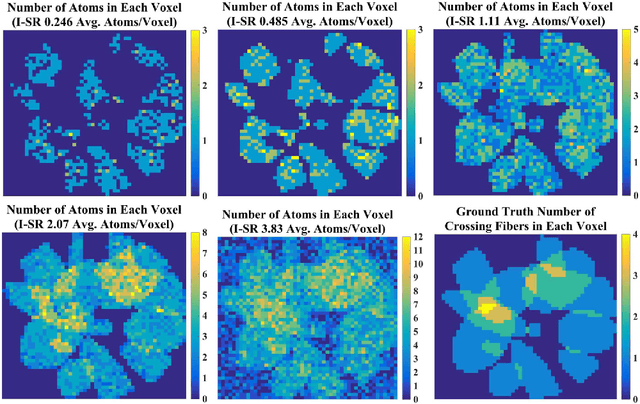
Abstract:Diffusion MRI (dMRI) provides the ability to reconstruct neuronal fibers in the brain, $\textit{in vivo}$, by measuring water diffusion along angular gradient directions in q-space. High angular resolution diffusion imaging (HARDI) can produce better estimates of fiber orientation than the popularly used diffusion tensor imaging, but the high number of samples needed to estimate diffusivity requires longer patient scan times. To accelerate dMRI, compressed sensing (CS) has been utilized by exploiting a sparse dictionary representation of the data, discovered through sparse coding. The sparser the representation, the fewer samples are needed to reconstruct a high resolution signal with limited information loss, and so an important area of research has focused on finding the sparsest possible representation of dMRI. Current reconstruction methods however, rely on an angular representation $\textit{per voxel}$ with added spatial regularization, and so, for non-zero signals, one is required to have at least one non-zero coefficient per voxel. This means that the global level of sparsity must be greater than the number of voxels. In contrast, we propose a joint spatial-angular representation of dMRI that will allow us to achieve levels of global sparsity that are below the number of voxels. A major challenge, however, is the computational complexity of solving a global sparse coding problem over large-scale dMRI. In this work, we present novel adaptations of popular sparse coding algorithms that become better suited for solving large-scale problems by exploiting spatial-angular separability. Our experiments show that our method achieves significantly sparser representations of HARDI than is possible by the state of the art.
(k,q)-Compressed Sensing for dMRI with Joint Spatial-Angular Sparsity Prior
Aug 02, 2017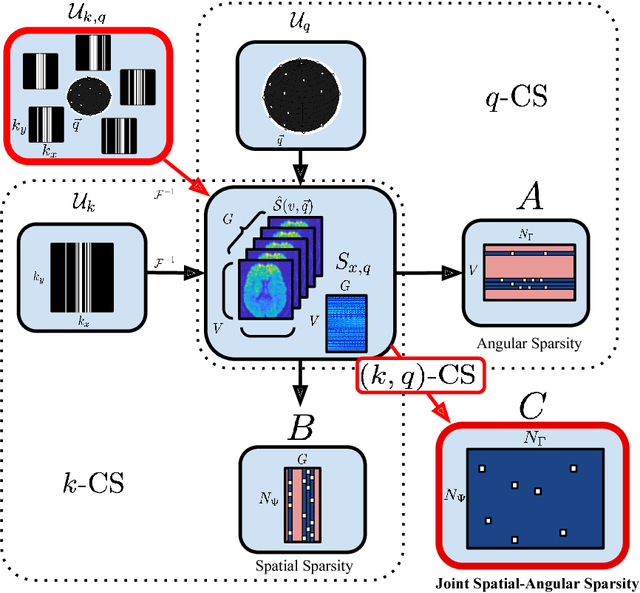
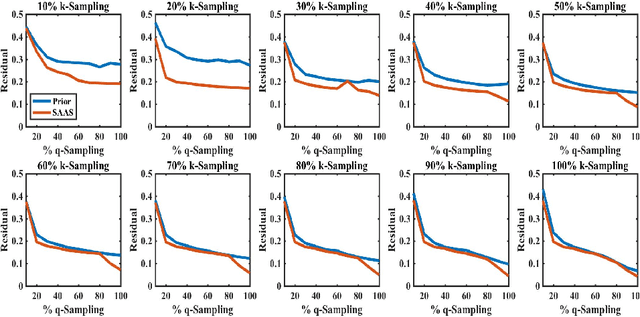
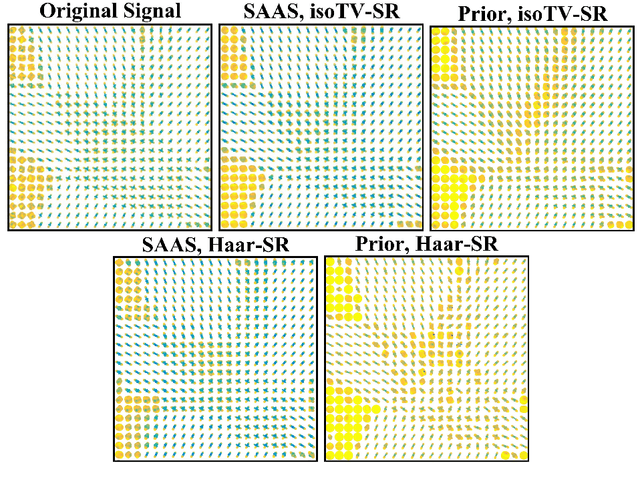
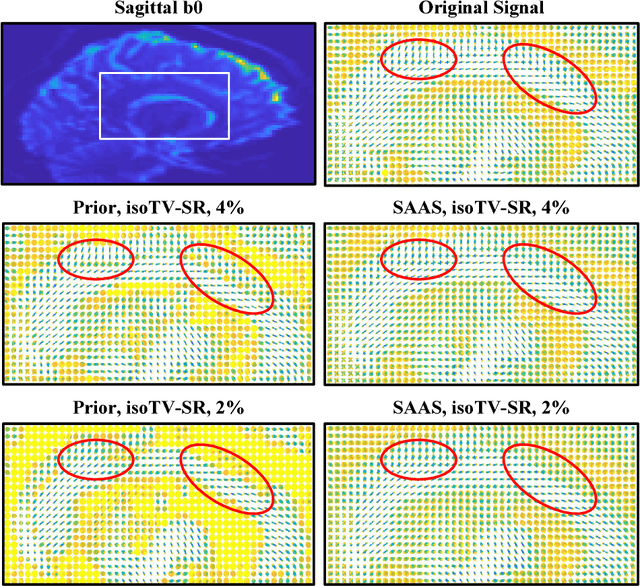
Abstract:Advanced diffusion magnetic resonance imaging (dMRI) techniques, like diffusion spectrum imaging (DSI) and high angular resolution diffusion imaging (HARDI), remain underutilized compared to diffusion tensor imaging because the scan times needed to produce accurate estimations of fiber orientation are significantly longer. To accelerate DSI and HARDI, recent methods from compressed sensing (CS) exploit a sparse underlying representation of the data in the spatial and angular domains to undersample in the respective k- and q-spaces. State-of-the-art frameworks, however, impose sparsity in the spatial and angular domains separately and involve the sum of the corresponding sparse regularizers. In contrast, we propose a unified (k,q)-CS formulation which imposes sparsity jointly in the spatial-angular domain to further increase sparsity of dMRI signals and reduce the required subsampling rate. To efficiently solve this large-scale global reconstruction problem, we introduce a novel adaptation of the FISTA algorithm that exploits dictionary separability. We show on phantom and real HARDI data that our approach achieves significantly more accurate signal reconstructions than the state of the art while sampling only 2-4% of the (k,q)-space, allowing for the potential of new levels of dMRI acceleration.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge