Enyu Cai
Semi-Supervised Object Detection for Sorghum Panicles in UAV Imagery
May 16, 2023



Abstract:The sorghum panicle is an important trait related to grain yield and plant development. Detecting and counting sorghum panicles can provide significant information for plant phenotyping. Current deep-learning-based object detection methods for panicles require a large amount of training data. The data labeling is time-consuming and not feasible for real application. In this paper, we present an approach to reduce the amount of training data for sorghum panicle detection via semi-supervised learning. Results show we can achieve similar performance as supervised methods for sorghum panicle detection by only using 10\% of original training data.
High-Resolution UAV Image Generation for Sorghum Panicle Detection
May 08, 2022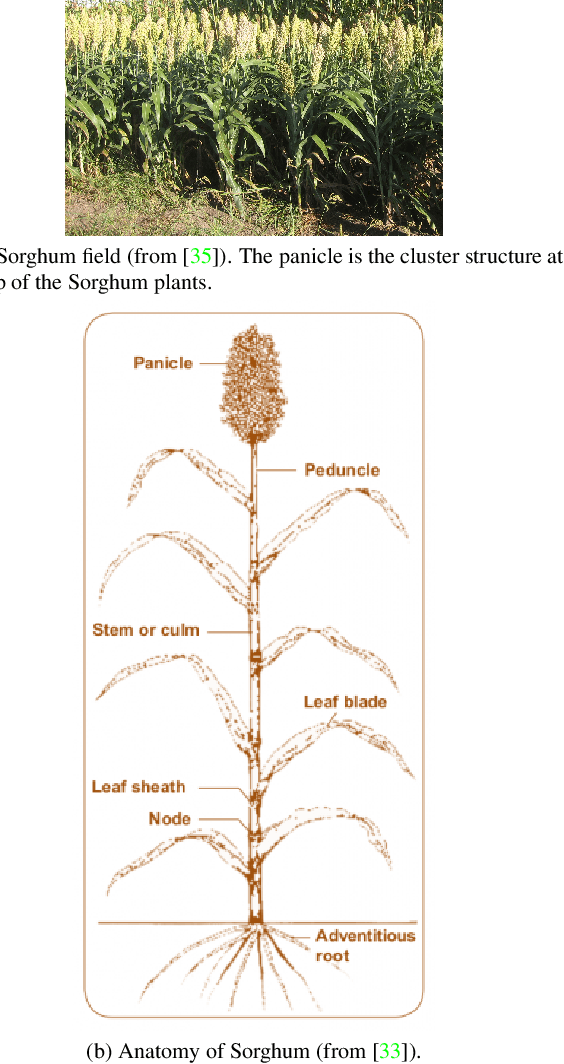
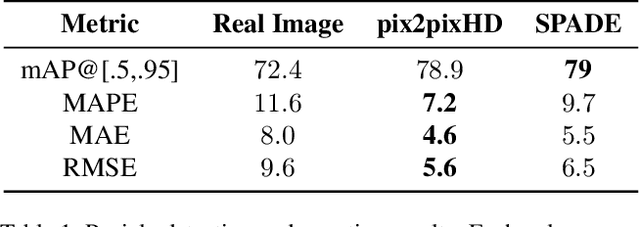

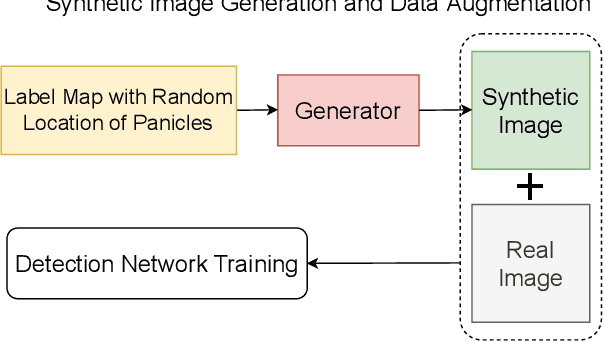
Abstract:The number of panicles (or heads) of Sorghum plants is an important phenotypic trait for plant development and grain yield estimation. The use of Unmanned Aerial Vehicles (UAVs) enables the capability of collecting and analyzing Sorghum images on a large scale. Deep learning can provide methods for estimating phenotypic traits from UAV images but requires a large amount of labeled data. The lack of training data due to the labor-intensive ground truthing of UAV images causes a major bottleneck in developing methods for Sorghum panicle detection and counting. In this paper, we present an approach that uses synthetic training images from generative adversarial networks (GANs) for data augmentation to enhance the performance of Sorghum panicle detection and counting. Our method can generate synthetic high-resolution UAV RGB images with panicle labels by using image-to-image translation GANs with a limited ground truth dataset of real UAV RGB images. The results show the improvements in panicle detection and counting using our data augmentation approach.
Field-Based Plot Extraction Using UAV RGB Images
Sep 01, 2021



Abstract:Unmanned Aerial Vehicles (UAVs) have become popular for use in plant phenotyping of field based crops, such as maize and sorghum, due to their ability to acquire high resolution data over field trials. Field experiments, which may comprise thousands of plants, are planted according to experimental designs to evaluate varieties or management practices. For many types of phenotyping analysis, we examine smaller groups of plants known as "plots." In this paper, we propose a new plot extraction method that will segment a UAV image into plots. We will demonstrate that our method achieves higher plot extraction accuracy than existing approaches.
Panicle Counting in UAV Images For Estimating Flowering Time in Sorghum
Jul 15, 2021



Abstract:Flowering time (time to flower after planting) is important for estimating plant development and grain yield for many crops including sorghum. Flowering time of sorghum can be approximated by counting the number of panicles (clusters of grains on a branch) across multiple dates. Traditional manual methods for panicle counting are time-consuming and tedious. In this paper, we propose a method for estimating flowering time and rapidly counting panicles using RGB images acquired by an Unmanned Aerial Vehicle (UAV). We evaluate three different deep neural network structures for panicle counting and location. Experimental results demonstrate that our method is able to accurately detect panicles and estimate sorghum flowering time.
Image-Based Plant Wilting Estimation
May 27, 2021



Abstract:Many plants become limp or droop through heat, loss of water, or disease. This is also known as wilting. In this paper, we examine plant wilting caused by bacterial infection. In particular, we want to design a metric for wilting based on images acquired of the plant. A quantifiable wilting metric will be useful in studying bacterial wilt and identifying resistance genes. Since there is no standard way to estimate wilting, it is common to use ad hoc visual scores. This is very subjective and requires expert knowledge of the plants and the disease mechanism. Our solution consists of using various wilting metrics acquired from RGB images of the plants. We also designed several experiments to demonstrate that our metrics are effective at estimating wilting in plants.
Deep Transfer Learning For Plant Center Localization
Apr 29, 2020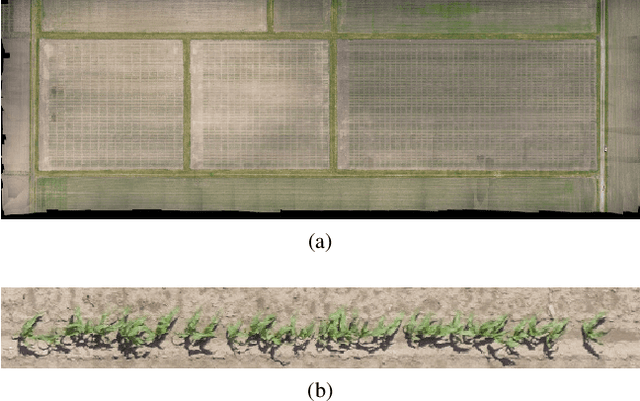



Abstract:Plant phenotyping focuses on the measurement of plant characteristics throughout the growing season, typically with the goal of evaluating genotypes for plant breeding. Estimating plant location is important for identifying genotypes which have low emergence, which is also related to the environment and management practices such as fertilizer applications. The goal of this paper is to investigate methods that estimate plant locations for a field-based crop using RGB aerial images captured using Unmanned Aerial Vehicles (UAVs). Deep learning approaches provide promising capability for locating plants observed in RGB images, but they require large quantities of labeled data (ground truth) for training. Using a deep learning architecture fine-tuned on a single field or a single type of crop on fields in other geographic areas or with other crops may not have good results. The problem of generating ground truth for each new field is labor-intensive and tedious. In this paper, we propose a method for estimating plant centers by transferring an existing model to a new scenario using limited ground truth data. We describe the use of transfer learning using a model fine-tuned for a single field or a single type of plant on a varied set of similar crops and fields. We show that transfer learning provides promising results for detecting plant locations.
Plant Stem Segmentation Using Fast Ground Truth Generation
Jan 24, 2020
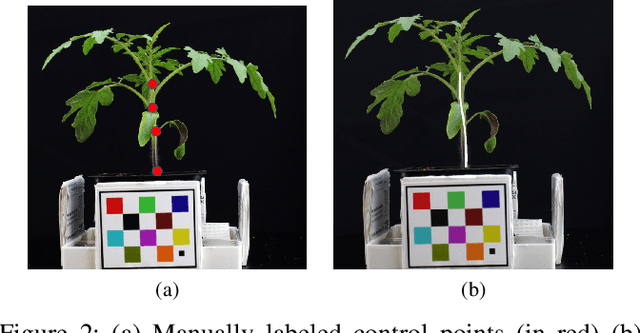

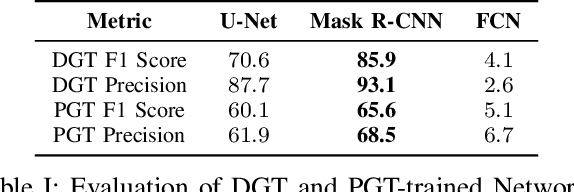
Abstract:Accurately phenotyping plant wilting is important for understanding responses to environmental stress. Analysis of the shape of plants can potentially be used to accurately quantify the degree of wilting. Plant shape analysis can be enhanced by locating the stem, which serves as a consistent reference point during wilting. In this paper, we show that deep learning methods can accurately segment tomato plant stems. We also propose a control-point-based ground truth method that drastically reduces the resources needed to create a training dataset for a deep learning approach. Experimental results show the viability of both our proposed ground truth approach and deep learning based stem segmentation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge