Emmanuelle Bourigault
UKBOB: One Billion MRI Labeled Masks for Generalizable 3D Medical Image Segmentation
Apr 09, 2025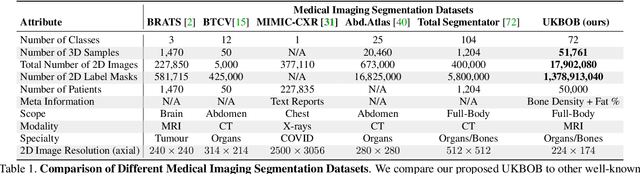
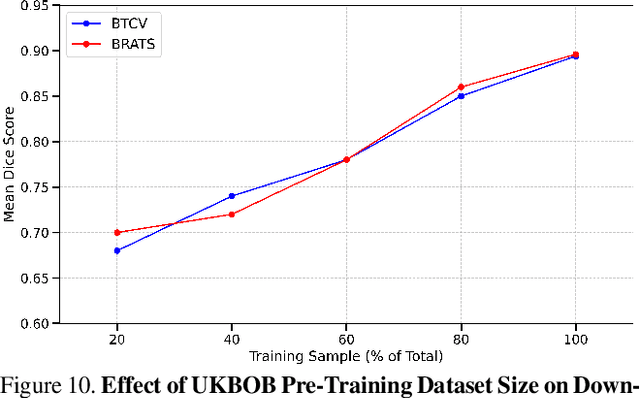
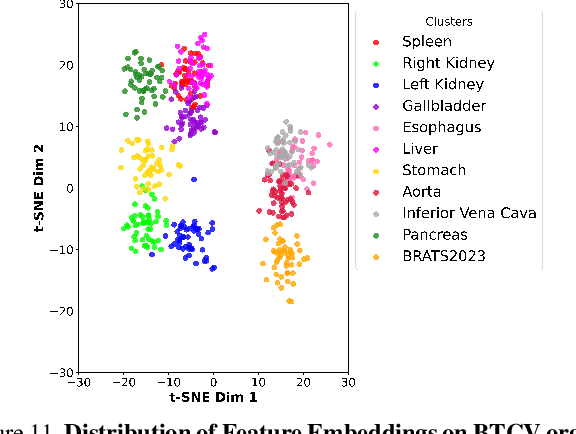
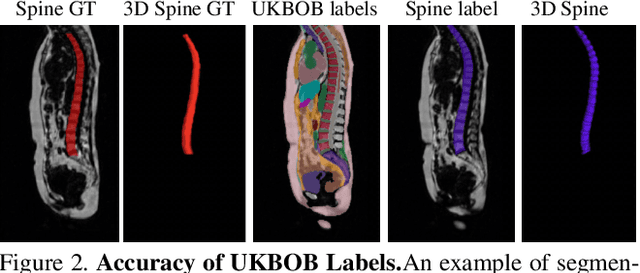
Abstract:In medical imaging, the primary challenge is collecting large-scale labeled data due to privacy concerns, logistics, and high labeling costs. In this work, we present the UK Biobank Organs and Bones (UKBOB), the largest labeled dataset of body organs, comprising 51,761 MRI 3D samples (equivalent to 17.9 million 2D images) and more than 1.37 billion 2D segmentation masks of 72 organs, all based on the UK Biobank MRI dataset. We utilize automatic labeling, introduce an automated label cleaning pipeline with organ-specific filters, and manually annotate a subset of 300 MRIs with 11 abdominal classes to validate the quality (referred to as UKBOB-manual). This approach allows for scaling up the dataset collection while maintaining confidence in the labels. We further confirm the validity of the labels by demonstrating zero-shot generalization of trained models on the filtered UKBOB to other small labeled datasets from similar domains (e.g., abdominal MRI). To further mitigate the effect of noisy labels, we propose a novel method called Entropy Test-time Adaptation (ETTA) to refine the segmentation output. We use UKBOB to train a foundation model, Swin-BOB, for 3D medical image segmentation based on the Swin-UNetr architecture, achieving state-of-the-art results in several benchmarks in 3D medical imaging, including the BRATS brain MRI tumor challenge (with a 0.4% improvement) and the BTCV abdominal CT scan benchmark (with a 1.3% improvement). The pre-trained models and the code are available at https://emmanuelleb985.github.io/ukbob , and the filtered labels will be made available with the UK Biobank.
3D Spine Shape Estimation from Single 2D DXA
Dec 02, 2024Abstract:Scoliosis is traditionally assessed based solely on 2D lateral deviations, but recent studies have also revealed the importance of other imaging planes in understanding the deformation of the spine. Consequently, extracting the spinal geometry in 3D would help quantify these spinal deformations and aid diagnosis. In this study, we propose an automated general framework to estimate the 3D spine shape from 2D DXA scans. We achieve this by explicitly predicting the sagittal view of the spine from the DXA scan. Using these two orthogonal projections of the spine (coronal in DXA, and sagittal from the prediction), we are able to describe the 3D shape of the spine. The prediction is learnt from over 30k paired images of DXA and MRI scans. We assess the performance of the method on a held out test set, and achieve high accuracy.
MVDiff: Scalable and Flexible Multi-View Diffusion for 3D Object Reconstruction from Single-View
May 06, 2024



Abstract:Generating consistent multiple views for 3D reconstruction tasks is still a challenge to existing image-to-3D diffusion models. Generally, incorporating 3D representations into diffusion model decrease the model's speed as well as generalizability and quality. This paper proposes a general framework to generate consistent multi-view images from single image or leveraging scene representation transformer and view-conditioned diffusion model. In the model, we introduce epipolar geometry constraints and multi-view attention to enforce 3D consistency. From as few as one image input, our model is able to generate 3D meshes surpassing baselines methods in evaluation metrics, including PSNR, SSIM and LPIPS.
X-Diffusion: Generating Detailed 3D MRI Volumes From a Single Image Using Cross-Sectional Diffusion Models
Apr 30, 2024



Abstract:In this work, we present X-Diffusion, a cross-sectional diffusion model tailored for Magnetic Resonance Imaging (MRI) data. X-Diffusion is capable of generating the entire MRI volume from just a single MRI slice or optionally from few multiple slices, setting new benchmarks in the precision of synthesized MRIs from extremely sparse observations. The uniqueness lies in the novel view-conditional training and inference of X-Diffusion on MRI volumes, allowing for generalized MRI learning. Our evaluations span both brain tumour MRIs from the BRATS dataset and full-body MRIs from the UK Biobank dataset. Utilizing the paired pre-registered Dual-energy X-ray Absorptiometry (DXA) and MRI modalities in the UK Biobank dataset, X-Diffusion is able to generate detailed 3D MRI volume from a single full-body DXA. Remarkably, the resultant MRIs not only stand out in precision on unseen examples (surpassing state-of-the-art results by large margins) but also flawlessly retain essential features of the original MRI, including tumour profiles, spine curvature, brain volume, and beyond. Furthermore, the trained X-Diffusion model on the MRI datasets attains a generalization capacity out-of-domain (e.g. generating knee MRIs even though it is trained on brains). The code is available on the project website https://emmanuelleb985.github.io/XDiffusion/ .
Multimodal PET/CT Tumour Segmentation and Prediction of Progression-Free Survival using a Full-Scale UNet with Attention
Nov 06, 2021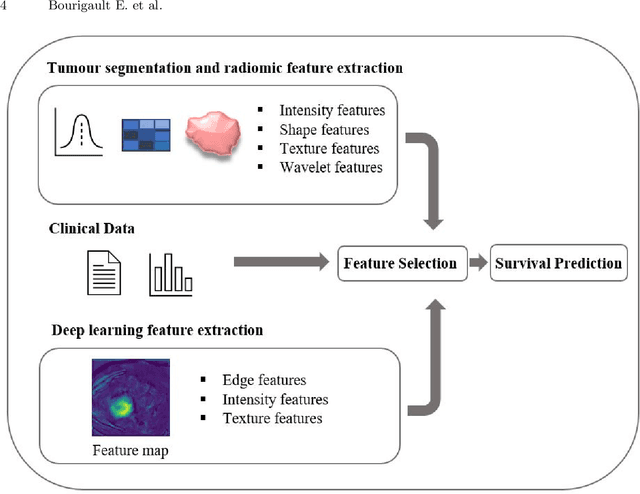
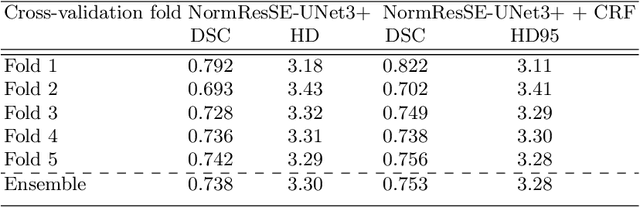

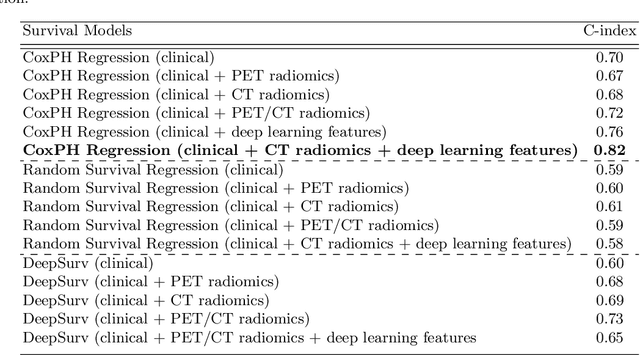
Abstract:Segmentation of head and neck (H\&N) tumours and prediction of patient outcome are crucial for patient's disease diagnosis and treatment monitoring. Current developments of robust deep learning models are hindered by the lack of large multi-centre, multi-modal data with quality annotations. The MICCAI 2021 HEad and neCK TumOR (HECKTOR) segmentation and outcome prediction challenge creates a platform for comparing segmentation methods of the primary gross target volume on fluoro-deoxyglucose (FDG)-PET and Computed Tomography images and prediction of progression-free survival in H\&N oropharyngeal cancer.For the segmentation task, we proposed a new network based on an encoder-decoder architecture with full inter- and intra-skip connections to take advantage of low-level and high-level semantics at full scales. Additionally, we used Conditional Random Fields as a post-processing step to refine the predicted segmentation maps. We trained multiple neural networks for tumor volume segmentation, and these segmentations were ensembled achieving an average Dice Similarity Coefficient of 0.75 in cross-validation, and 0.76 on the challenge testing data set. For prediction of patient progression free survival task, we propose a Cox proportional hazard regression combining clinical, radiomic, and deep learning features. Our survival prediction model achieved a concordance index of 0.82 in cross-validation, and 0.62 on the challenge testing data set.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge