Amir Jamaludin
UKBOB: One Billion MRI Labeled Masks for Generalizable 3D Medical Image Segmentation
Apr 09, 2025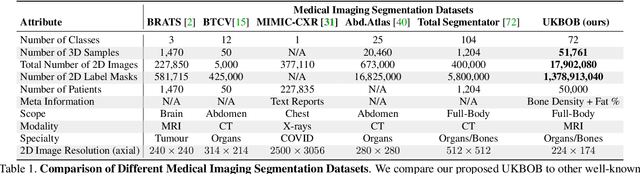
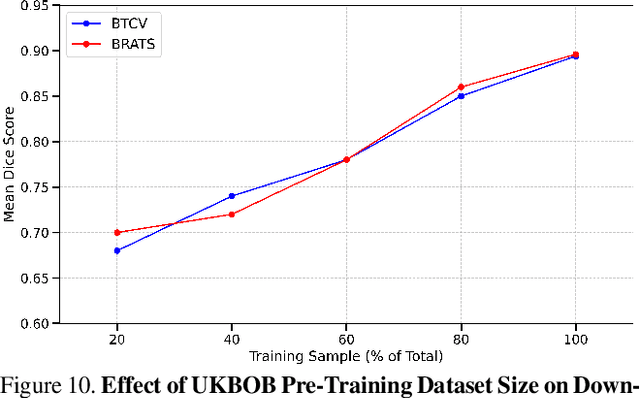
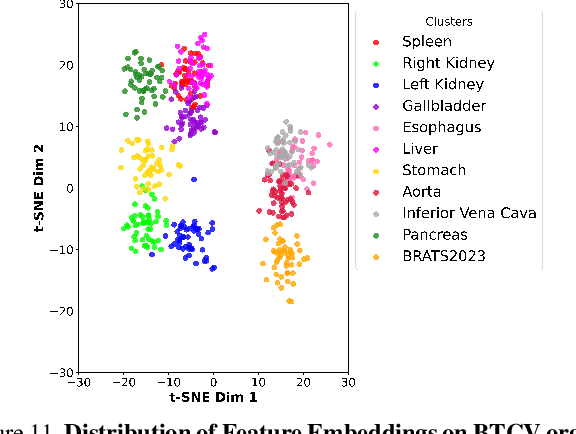
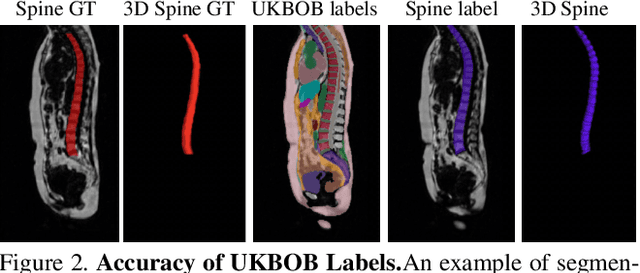
Abstract:In medical imaging, the primary challenge is collecting large-scale labeled data due to privacy concerns, logistics, and high labeling costs. In this work, we present the UK Biobank Organs and Bones (UKBOB), the largest labeled dataset of body organs, comprising 51,761 MRI 3D samples (equivalent to 17.9 million 2D images) and more than 1.37 billion 2D segmentation masks of 72 organs, all based on the UK Biobank MRI dataset. We utilize automatic labeling, introduce an automated label cleaning pipeline with organ-specific filters, and manually annotate a subset of 300 MRIs with 11 abdominal classes to validate the quality (referred to as UKBOB-manual). This approach allows for scaling up the dataset collection while maintaining confidence in the labels. We further confirm the validity of the labels by demonstrating zero-shot generalization of trained models on the filtered UKBOB to other small labeled datasets from similar domains (e.g., abdominal MRI). To further mitigate the effect of noisy labels, we propose a novel method called Entropy Test-time Adaptation (ETTA) to refine the segmentation output. We use UKBOB to train a foundation model, Swin-BOB, for 3D medical image segmentation based on the Swin-UNetr architecture, achieving state-of-the-art results in several benchmarks in 3D medical imaging, including the BRATS brain MRI tumor challenge (with a 0.4% improvement) and the BTCV abdominal CT scan benchmark (with a 1.3% improvement). The pre-trained models and the code are available at https://emmanuelleb985.github.io/ukbob , and the filtered labels will be made available with the UK Biobank.
3D Spine Shape Estimation from Single 2D DXA
Dec 02, 2024Abstract:Scoliosis is traditionally assessed based solely on 2D lateral deviations, but recent studies have also revealed the importance of other imaging planes in understanding the deformation of the spine. Consequently, extracting the spinal geometry in 3D would help quantify these spinal deformations and aid diagnosis. In this study, we propose an automated general framework to estimate the 3D spine shape from 2D DXA scans. We achieve this by explicitly predicting the sagittal view of the spine from the DXA scan. Using these two orthogonal projections of the spine (coronal in DXA, and sagittal from the prediction), we are able to describe the 3D shape of the spine. The prediction is learnt from over 30k paired images of DXA and MRI scans. We assess the performance of the method on a held out test set, and achieve high accuracy.
Automated Spinal MRI Labelling from Reports Using a Large Language Model
Oct 22, 2024Abstract:We propose a general pipeline to automate the extraction of labels from radiology reports using large language models, which we validate on spinal MRI reports. The efficacy of our labelling method is measured on five distinct conditions: spinal cancer, stenosis, spondylolisthesis, cauda equina compression and herniation. Using open-source models, our method equals or surpasses GPT-4 on a held-out set of reports. Furthermore, we show that the extracted labels can be used to train imaging models to classify the identified conditions in the accompanying MR scans. All classifiers trained using automated labels achieve comparable performance to models trained using scans manually annotated by clinicians. Code can be found at https://github.com/robinyjpark/AutoLabelClassifier.
X-Diffusion: Generating Detailed 3D MRI Volumes From a Single Image Using Cross-Sectional Diffusion Models
Apr 30, 2024



Abstract:In this work, we present X-Diffusion, a cross-sectional diffusion model tailored for Magnetic Resonance Imaging (MRI) data. X-Diffusion is capable of generating the entire MRI volume from just a single MRI slice or optionally from few multiple slices, setting new benchmarks in the precision of synthesized MRIs from extremely sparse observations. The uniqueness lies in the novel view-conditional training and inference of X-Diffusion on MRI volumes, allowing for generalized MRI learning. Our evaluations span both brain tumour MRIs from the BRATS dataset and full-body MRIs from the UK Biobank dataset. Utilizing the paired pre-registered Dual-energy X-ray Absorptiometry (DXA) and MRI modalities in the UK Biobank dataset, X-Diffusion is able to generate detailed 3D MRI volume from a single full-body DXA. Remarkably, the resultant MRIs not only stand out in precision on unseen examples (surpassing state-of-the-art results by large margins) but also flawlessly retain essential features of the original MRI, including tumour profiles, spine curvature, brain volume, and beyond. Furthermore, the trained X-Diffusion model on the MRI datasets attains a generalization capacity out-of-domain (e.g. generating knee MRIs even though it is trained on brains). The code is available on the project website https://emmanuelleb985.github.io/XDiffusion/ .
Predicting Spine Geometry and Scoliosis from DXA Scans
Nov 15, 2023



Abstract:Our objective in this paper is to estimate spine curvature in DXA scans. To this end we first train a neural network to predict the middle spine curve in the scan, and then use an integral-based method to determine the curvature along the spine curve. We use the curvature to compare to the standard angle scoliosis measure obtained using the DXA Scoliosis Method (DSM). The performance improves over the prior work of Jamaludin et al. 2018. We show that the maximum curvature can be used as a scoring function for ordering the severity of spinal deformation.
Contouring by Unit Vector Field Regression
May 26, 2023



Abstract:This work introduces a simple deep-learning based method to delineate contours by `walking' along learnt unit vector fields. We demonstrate the effectiveness of our pipeline on the unique case of open contours on the task of delineating the sacroiliac joints (SIJs) in spinal MRIs. We show that: (i) 95% of the time the average root mean square error of the predicted contour against the original ground truth is below 4.5 pixels (2.5mm for a standard T1-weighted SIJ MRI), and (ii) the proposed method is better than the baseline of regressing vertices or landmarks of contours.
Vision-Language Modelling For Radiological Imaging and Reports In The Low Data Regime
Mar 30, 2023



Abstract:This paper explores training medical vision-language models (VLMs) -- where the visual and language inputs are embedded into a common space -- with a particular focus on scenarios where training data is limited, as is often the case in clinical datasets. We explore several candidate methods to improve low-data performance, including: (i) adapting generic pre-trained models to novel image and text domains (i.e. medical imaging and reports) via unimodal self-supervision; (ii) using local (e.g. GLoRIA) & global (e.g. InfoNCE) contrastive loss functions as well as a combination of the two; (iii) extra supervision during VLM training, via: (a) image- and text-only self-supervision, and (b) creating additional positive image-text pairs for training through augmentation and nearest-neighbour search. Using text-to-image retrieval as a benchmark, we evaluate the performance of these methods with variable sized training datasets of paired chest X-rays and radiological reports. Combined, they significantly improve retrieval compared to fine-tuning CLIP, roughly equivalent to training with the data. A similar pattern is found in the downstream task classification of CXR-related conditions with our method outperforming CLIP and also BioVIL, a strong CXR VLM benchmark, in the zero-shot and linear probing settings. We conclude with a set of recommendations for researchers aiming to train vision-language models on other medical imaging modalities when training data is scarce. To facilitate further research, we will make our code and models publicly available.
Context-Aware Transformers For Spinal Cancer Detection and Radiological Grading
Jun 27, 2022



Abstract:This paper proposes a novel transformer-based model architecture for medical imaging problems involving analysis of vertebrae. It considers two applications of such models in MR images: (a) detection of spinal metastases and the related conditions of vertebral fractures and metastatic cord compression, (b) radiological grading of common degenerative changes in intervertebral discs. Our contributions are as follows: (i) We propose a Spinal Context Transformer (SCT), a deep-learning architecture suited for the analysis of repeated anatomical structures in medical imaging such as vertebral bodies (VBs). Unlike previous related methods, SCT considers all VBs as viewed in all available image modalities together, making predictions for each based on context from the rest of the spinal column and all available imaging modalities. (ii) We apply the architecture to a novel and important task: detecting spinal metastases and the related conditions of cord compression and vertebral fractures/collapse from multi-series spinal MR scans. This is done using annotations extracted from free-text radiological reports as opposed to bespoke annotation. However, the resulting model shows strong agreement with vertebral-level bespoke radiologist annotations on the test set. (iii) We also apply SCT to an existing problem: radiological grading of inter-vertebral discs (IVDs) in lumbar MR scans for common degenerative changes.We show that by considering the context of vertebral bodies in the image, SCT improves the accuracy for several gradings compared to previously published model.
SpineNetV2: Automated Detection, Labelling and Radiological Grading Of Clinical MR Scans
May 03, 2022



Abstract:This technical report presents SpineNetV2, an automated tool which: (i) detects and labels vertebral bodies in clinical spinal magnetic resonance (MR) scans across a range of commonly used sequences; and (ii) performs radiological grading of lumbar intervertebral discs in T2-weighted scans for a range of common degenerative changes. SpineNetV2 improves over the original SpineNet software in two ways: (1) The vertebral body detection stage is significantly faster, more accurate and works across a range of fields-of-view (as opposed to just lumbar scans). (2) Radiological grading adopts a more powerful architecture, adding several new grading schemes without loss in performance. A demo of the software is available at the project website: http://zeus.robots.ox.ac.uk/spinenet2/.
Self-Supervised Multi-Modal Alignment for Whole Body Medical Imaging
Aug 06, 2021



Abstract:This paper explores the use of self-supervised deep learning in medical imaging in cases where two scan modalities are available for the same subject. Specifically, we use a large publicly-available dataset of over 20,000 subjects from the UK Biobank with both whole body Dixon technique magnetic resonance (MR) scans and also dual-energy x-ray absorptiometry (DXA) scans. We make three contributions: (i) We introduce a multi-modal image-matching contrastive framework, that is able to learn to match different-modality scans of the same subject with high accuracy. (ii) Without any adaption, we show that the correspondences learnt during this contrastive training step can be used to perform automatic cross-modal scan registration in a completely unsupervised manner. (iii) Finally, we use these registrations to transfer segmentation maps from the DXA scans to the MR scans where they are used to train a network to segment anatomical regions without requiring ground-truth MR examples. To aid further research, our code will be made publicly available.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge