Elisabeth Ailer
Targeted Sequential Indirect Experiment Design
May 30, 2024Abstract:Scientific hypotheses typically concern specific aspects of complex, imperfectly understood or entirely unknown mechanisms, such as the effect of gene expression levels on phenotypes or how microbial communities influence environmental health. Such queries are inherently causal (rather than purely associational), but in many settings, experiments can not be conducted directly on the target variables of interest, but are indirect. Therefore, they perturb the target variable, but do not remove potential confounding factors. If, additionally, the resulting experimental measurements are multi-dimensional and the studied mechanisms nonlinear, the query of interest is generally not identified. We develop an adaptive strategy to design indirect experiments that optimally inform a targeted query about the ground truth mechanism in terms of sequentially narrowing the gap between an upper and lower bound on the query. While the general formulation consists of a bi-level optimization procedure, we derive an efficiently estimable analytical kernel-based estimator of the bounds for the causal effect, a query of key interest, and demonstrate the efficacy of our approach in confounded, multivariate, nonlinear synthetic settings.
Sequential Underspecified Instrument Selection for Cause-Effect Estimation
Feb 11, 2023



Abstract:Instrumental variable (IV) methods are used to estimate causal effects in settings with unobserved confounding, where we cannot directly experiment on the treatment variable. Instruments are variables which only affect the outcome indirectly via the treatment variable(s). Most IV applications focus on low-dimensional treatments and crucially require at least as many instruments as treatments. This assumption is restrictive: in the natural sciences we often seek to infer causal effects of high-dimensional treatments (e.g., the effect of gene expressions or microbiota on health and disease), but can only run few experiments with a limited number of instruments (e.g., drugs or antibiotics). In such underspecified problems, the full treatment effect is not identifiable in a single experiment even in the linear case. We show that one can still reliably recover the projection of the treatment effect onto the instrumented subspace and develop techniques to consistently combine such partial estimates from different sets of instruments. We then leverage our combined estimators in an algorithm that iteratively proposes the most informative instruments at each round of experimentation to maximize the overall information about the full causal effect.
Supervised Learning and Model Analysis with Compositional Data
May 15, 2022



Abstract:The compositionality and sparsity of high-throughput sequencing data poses a challenge for regression and classification. However, in microbiome research in particular, conditional modeling is an essential tool to investigate relationships between phenotypes and the microbiome. Existing techniques are often inadequate: they either rely on extensions of the linear log-contrast model (which adjusts for compositionality, but is often unable to capture useful signals), or they are based on black-box machine learning methods (which may capture useful signals, but ignore compositionality in downstream analyses). We propose KernelBiome, a kernel-based nonparametric regression and classification framework for compositional data. It is tailored to sparse compositional data and is able to incorporate prior knowledge, such as phylogenetic structure. KernelBiome captures complex signals, including in the zero-structure, while automatically adapting model complexity. We demonstrate on par or improved predictive performance compared with state-of-the-art machine learning methods. Additionally, our framework provides two key advantages: (i) We propose two novel quantities to interpret contributions of individual components and prove that they consistently estimate average perturbation effects of the conditional mean, extending the interpretability of linear log-contrast models to nonparametric models. (ii) We show that the connection between kernels and distances aids interpretability and provides a data-driven embedding that can augment further analysis. Finally, we apply the KernelBiome framework to two public microbiome studies and illustrate the proposed model analysis. KernelBiome is available as an open-source Python package at https://github.com/shimenghuang/KernelBiome.
A causal view on compositional data
Jun 21, 2021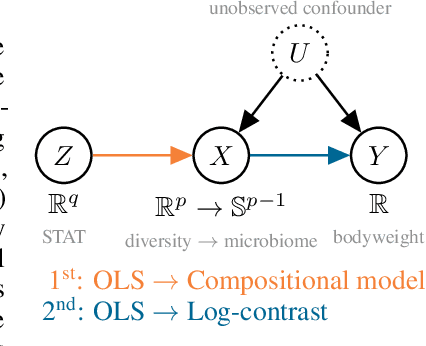
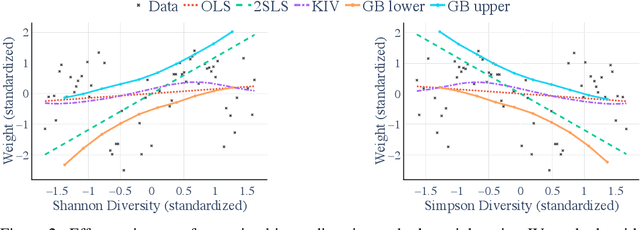
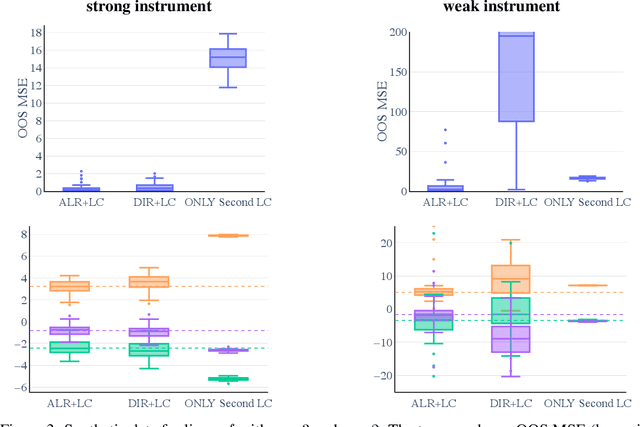
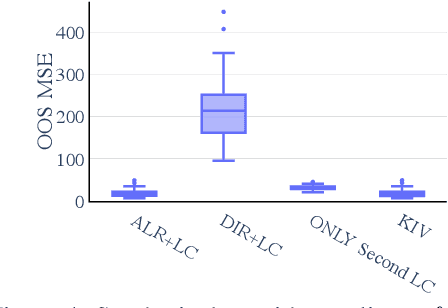
Abstract:Many scientific datasets are compositional in nature. Important examples include species abundances in ecology, rock compositions in geology, topic compositions in large-scale text corpora, and sequencing count data in molecular biology. Here, we provide a causal view on compositional data in an instrumental variable setting where the composition acts as the cause. Throughout, we pay particular attention to the interpretation of compositional causes from the viewpoint of interventions and crisply articulate potential pitfalls for practitioners. Focusing on modern high-dimensional microbiome sequencing data as a timely illustrative use case, our analysis first reveals that popular one-dimensional information-theoretic summary statistics, such as diversity and richness, may be insufficient for drawing causal conclusions from ecological data. Instead, we advocate for multivariate alternatives using statistical data transformations and regression techniques that take the special structure of the compositional sample space into account. In a comparative analysis on synthetic and semi-synthetic data we show the advantages and limitations of our proposal. We posit that our framework may provide a useful starting point for cause-effect estimation in the context of compositional data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge