Edmond Cunningham
Principal Manifold Flows
Feb 14, 2022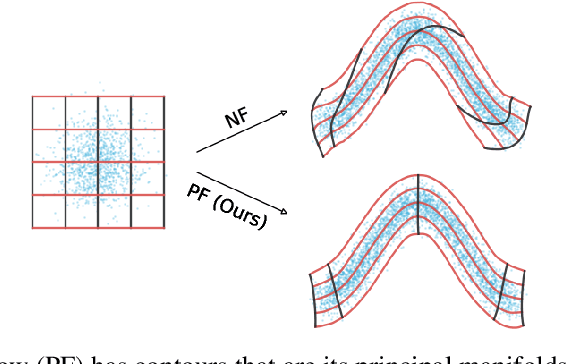
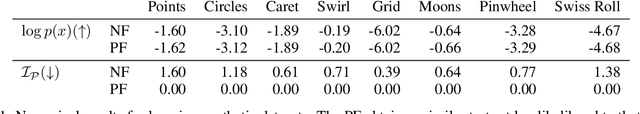
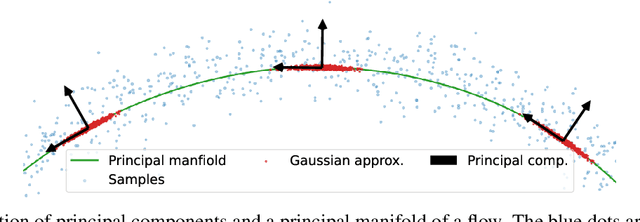

Abstract:Normalizing flows map an independent set of latent variables to their samples using a bijective transformation. Despite the exact correspondence between samples and latent variables, their high level relationship is not well understood. In this paper we characterize the geometric structure of flows using principal manifolds and understand the relationship between latent variables and samples using contours. We introduce a novel class of normalizing flows, called principal manifold flows (PF), whose contours are its principal manifolds, and a variant for injective flows (iPF) that is more efficient to train than regular injective flows. PFs can be constructed using any flow architecture, are trained with a regularized maximum likelihood objective and can perform density estimation on all of their principal manifolds. In our experiments we show that PFs and iPFs are able to learn the principal manifolds over a variety of datasets. Additionally, we show that PFs can perform density estimation on data that lie on a manifold with variable dimensionality, which is not possible with existing normalizing flows.
Normalizing Flows Across Dimensions
Jun 23, 2020
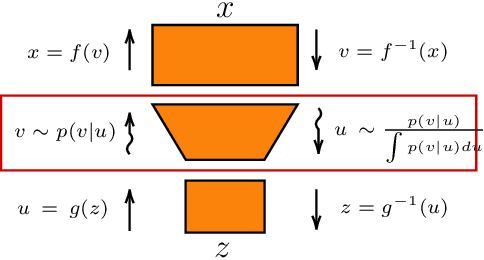
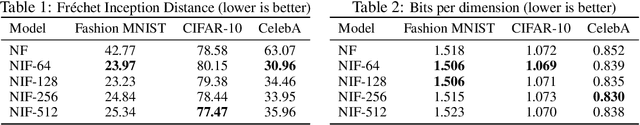
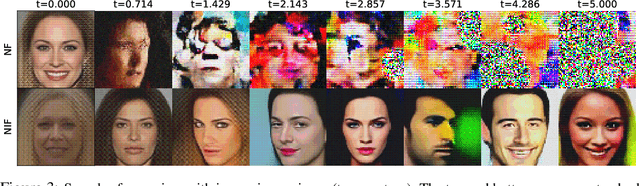
Abstract:Real-world data with underlying structure, such as pictures of faces, are hypothesized to lie on a low-dimensional manifold. This manifold hypothesis has motivated state-of-the-art generative algorithms that learn low-dimensional data representations. Unfortunately, a popular generative model, normalizing flows, cannot take advantage of this. Normalizing flows are based on successive variable transformations that are, by design, incapable of learning lower-dimensional representations. In this paper we introduce noisy injective flows (NIF), a generalization of normalizing flows that can go across dimensions. NIF explicitly map the latent space to a learnable manifold in a high-dimensional data space using injective transformations. We further employ an additive noise model to account for deviations from the manifold and identify a stochastic inverse of the generative process. Empirically, we demonstrate that a simple application of our method to existing flow architectures can significantly improve sample quality and yield separable data embeddings.
Explainable Genetic Inheritance Pattern Prediction
Dec 05, 2018
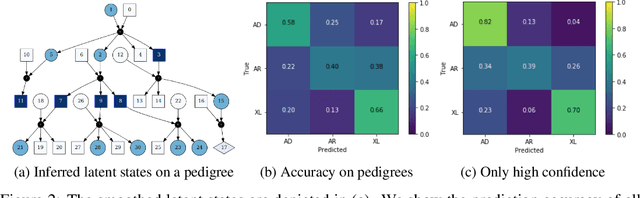
Abstract:Diagnosing an inherited disease often requires identifying the pattern of inheritance in a patient's family. We represent family trees with genetic patterns of inheritance using hypergraphs and latent state space models to provide explainable inheritance pattern predictions. Our approach allows for exact causal inference over a patient's possible genotypes given their relatives' phenotypes. By design, inference can be examined at a low level to provide explainable predictions. Furthermore, we make use of human intuition by providing a method to assign hypothetical evidence to any inherited gene alleles. Our analysis supports the application of latent state space models to improve patient care in cases of rare inherited diseases where access to genetic specialists is limited.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge