Duowen Chen
Trajectory Consistency for One-Step Generation on Euler Mean Flows
Jan 31, 2026Abstract:We propose \emph{Euler Mean Flows (EMF)}, a flow-based generative framework for one-step and few-step generation that enforces long-range trajectory consistency with minimal sampling cost. The key idea of EMF is to replace the trajectory consistency constraint, which is difficult to supervise and optimize over long time scales, with a principled linear surrogate that enables direct data supervision for long-horizon flow-map compositions. We derive this approximation from the semigroup formulation of flow-based models and show that, under mild regularity assumptions, it faithfully approximates the original consistency objective while being substantially easier to optimize. This formulation leads to a unified, JVP-free training framework that supports both $u$-prediction and $x_1$-prediction variants, avoiding explicit Jacobian computations and significantly reducing memory and computational overhead. Experiments on image synthesis, particle-based geometry generation, and functional generation demonstrate improved optimization stability and sample quality under fixed sampling budgets, together with approximately $50\%$ reductions in training time and memory consumption compared to existing one-step methods for image generation.
Bidirectional Copy-Paste for Semi-Supervised Medical Image Segmentation
May 01, 2023Abstract:In semi-supervised medical image segmentation, there exist empirical mismatch problems between labeled and unlabeled data distribution. The knowledge learned from the labeled data may be largely discarded if treating labeled and unlabeled data separately or in an inconsistent manner. We propose a straightforward method for alleviating the problem - copy-pasting labeled and unlabeled data bidirectionally, in a simple Mean Teacher architecture. The method encourages unlabeled data to learn comprehensive common semantics from the labeled data in both inward and outward directions. More importantly, the consistent learning procedure for labeled and unlabeled data can largely reduce the empirical distribution gap. In detail, we copy-paste a random crop from a labeled image (foreground) onto an unlabeled image (background) and an unlabeled image (foreground) onto a labeled image (background), respectively. The two mixed images are fed into a Student network and supervised by the mixed supervisory signals of pseudo-labels and ground-truth. We reveal that the simple mechanism of copy-pasting bidirectionally between labeled and unlabeled data is good enough and the experiments show solid gains (e.g., over 21% Dice improvement on ACDC dataset with 5% labeled data) compared with other state-of-the-arts on various semi-supervised medical image segmentation datasets. Code is available at https://github.com/DeepMed-Lab-ECNU/BCP}.
MagicNet: Semi-Supervised Multi-Organ Segmentation via Magic-Cube Partition and Recovery
Dec 29, 2022Abstract:We propose a novel teacher-student model for semi-supervised multi-organ segmentation. In teacher-student model, data augmentation is usually adopted on unlabeled data to regularize the consistent training between teacher and student. We start from a key perspective that fixed relative locations and variable sizes of different organs can provide distribution information where a multi-organ CT scan is drawn. Thus, we treat the prior anatomy as a strong tool to guide the data augmentation and reduce the mismatch between labeled and unlabeled images for semi-supervised learning. More specifically, we propose a data augmentation strategy based on partition-and-recovery N$^3$ cubes cross- and within- labeled and unlabeled images. Our strategy encourages unlabeled images to learn organ semantics in relative locations from the labeled images (cross-branch) and enhances the learning ability for small organs (within-branch). For within-branch, we further propose to refine the quality of pseudo labels by blending the learned representations from small cubes to incorporate local attributes. Our method is termed as MagicNet, since it treats the CT volume as a magic-cube and $N^3$-cube partition-and-recovery process matches with the rule of playing a magic-cube. Extensive experiments on two public CT multi-organ datasets demonstrate the effectiveness of MagicNet, and noticeably outperforms state-of-the-art semi-supervised medical image segmentation approaches, with +7% DSC improvement on MACT dataset with 10% labeled images.
SB-GCN: Structured BREP Graph Convolutional Network for Automatic Mating of CAD Assemblies
May 25, 2021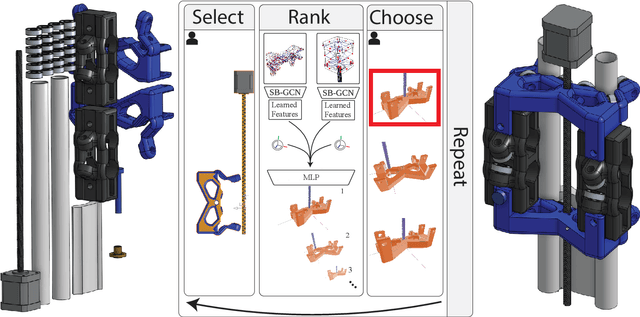
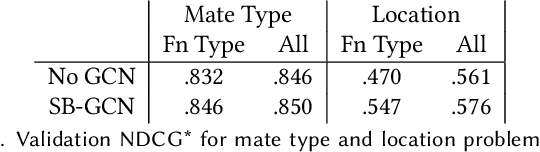

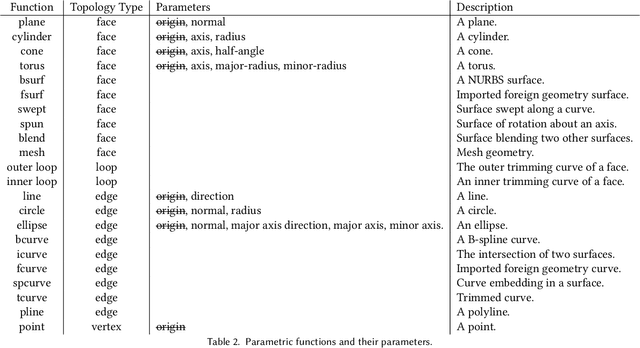
Abstract:Assembly modeling is a core task of computer aided design (CAD), comprising around one third of the work in a CAD workflow. Optimizing this process therefore represents a huge opportunity in the design of a CAD system, but current research of assembly based modeling is not directly applicable to modern CAD systems because it eschews the dominant data structure of modern CAD: parametric boundary representations (BREPs). CAD assembly modeling defines assemblies as a system of pairwise constraints, called mates, between parts, which are defined relative to BREP topology rather than in world coordinates common to existing work. We propose SB-GCN, a representation learning scheme on BREPs that retains the topological structure of parts, and use these learned representations to predict CAD type mates. To train our system, we compiled the first large scale dataset of BREP CAD assemblies, which we are releasing along with benchmark mate prediction tasks. Finally, we demonstrate the compatibility of our model with an existing commercial CAD system by building a tool that assists users in mate creation by suggesting mate completions, with 72.2% accuracy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge