Dorothee Auer
A Unified Framework for Semi-Supervised Image Segmentation and Registration
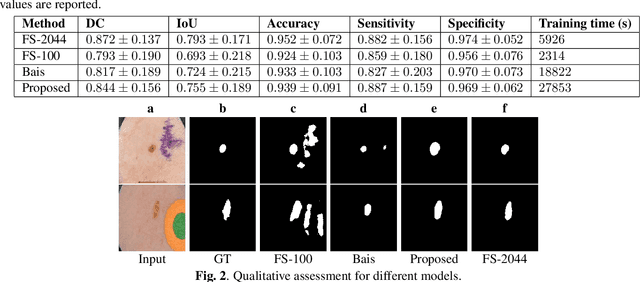
Feb 05, 2025Abstract:Semi-supervised learning, which leverages both annotated and unannotated data, is an efficient approach for medical image segmentation, where obtaining annotations for the whole dataset is time-consuming and costly. Traditional semi-supervised methods primarily focus on extracting features and learning data distributions from unannotated data to enhance model training. In this paper, we introduce a novel approach incorporating an image registration model to generate pseudo-labels for the unannotated data, producing more geometrically correct pseudo-labels to improve the model training. Our method was evaluated on a 2D brain data set, showing excellent performance even using only 1\% of the annotated data. The results show that our approach outperforms conventional semi-supervised segmentation methods (e.g. teacher-student model), particularly in a low percentage of annotation scenario. GitHub: https://github.com/ruizhe-l/UniSegReg.
Pinv-Recon: Generalized MR Image Reconstruction via Pseudoinversion of the Encoding Matrix
Oct 08, 2024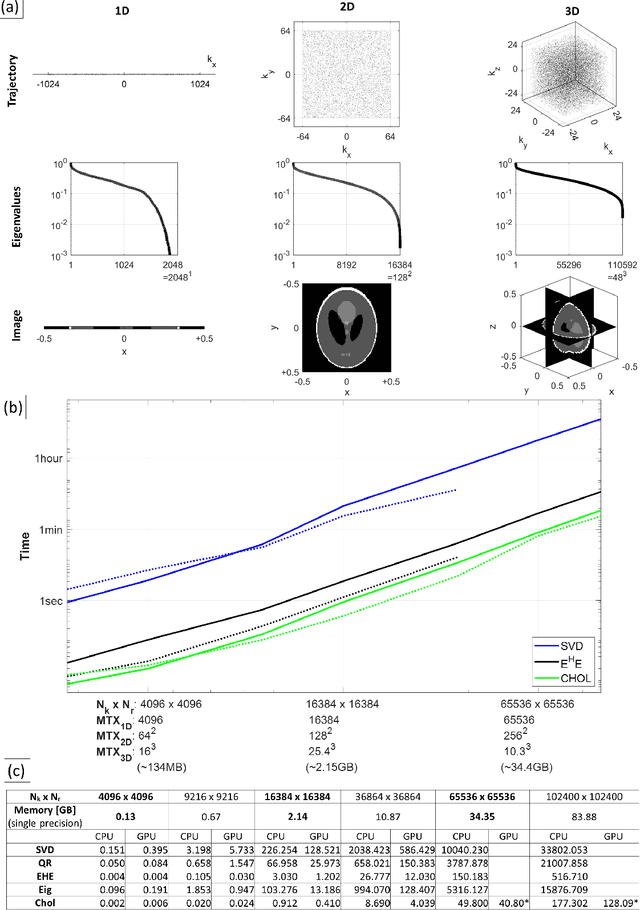
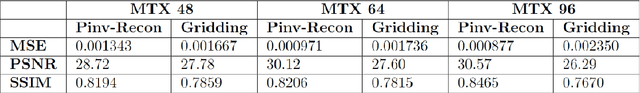
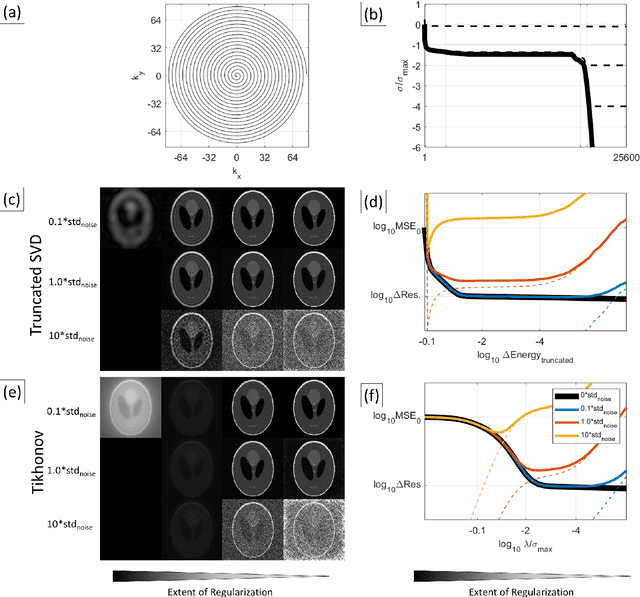
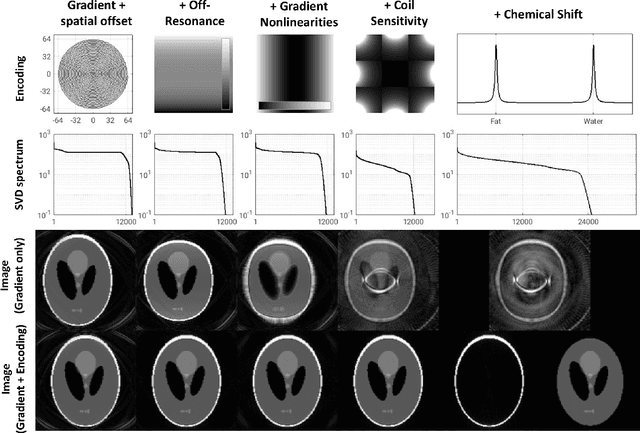
Abstract:Purpose: To present a novel generalized MR image reconstruction based on pseudoinversion of the encoding matrix (Pinv-Recon) as a simple yet powerful method, and demonstrate its computational feasibility for diverse MR imaging applications. Methods: MR image encoding constitutes a linear mapping of the unknown image to the measured k-space data mediated via an encoding matrix ($ data = Encode \times image$). Pinv-Recon addresses MR image reconstruction as a linear inverse problem ($image = Encode^{-1} \times data$), explicitly calculating the Moore-Penrose pseudoinverse of the encoding matrix using truncated singular value decomposition (tSVD). Using a discretized, algebraic notation, we demonstrate constructing a generalized encoding matrix by stacking relevant encoding mechanisms (e.g., gradient encoding, coil sensitivity encoding, chemical shift inversion) and encoding distortions (e.g., off-center positioning, B$_0$ inhomogeneity, spatiotemporal gradient imperfections, transient relaxation effects). Iterative reconstructions using the explicit generalized encoding matrix, and the computation of the spatial-response-function (SRF) and noise amplification, were demonstrated. Results: We evaluated the computation times and memory requirements (time ~ (size of the encoding matrix)$^{1.4}$). Using the Shepp-Logan phantom, we demonstrated the versatility of the method for various intertwined MR image encoding and distortion mechanisms, achieving better MSE, PSNR and SSIM metrics than conventional methods. A diversity of datasets, including the ISMRM CG-SENSE challenge, were used to validate Pinv-Recon. Conclusion: Although pseudo-inversion of large encoding matrices was once deemed computationally intractable, recent advances make Pinv-Recon feasible. It has great promise for both research and clinical applications, and for educational use.
MrRegNet: Multi-resolution Mask Guided Convolutional Neural Network for Medical Image Registration with Large Deformations
May 16, 2024



Abstract:Deformable image registration (alignment) is highly sought after in numerous clinical applications, such as computer aided diagnosis and disease progression analysis. Deep Convolutional Neural Network (DCNN)-based image registration methods have demonstrated advantages in terms of registration accuracy and computational speed. However, while most methods excel at global alignment, they often perform worse in aligning local regions. To address this challenge, this paper proposes a mask-guided encoder-decoder DCNN-based image registration method, named as MrRegNet. This approach employs a multi-resolution encoder for feature extraction and subsequently estimates multi-resolution displacement fields in the decoder to handle the substantial deformation of images. Furthermore, segmentation masks are employed to direct the model's attention toward aligning local regions. The results show that the proposed method outperforms traditional methods like Demons and a well-known deep learning method, VoxelMorph, on a public 3D brain MRI dataset (OASIS) and a local 2D brain MRI dataset with large deformations. Importantly, the image alignment accuracies are significantly improved at local regions guided by segmentation masks. Github link:https://github.com/ruizhe-l/MrRegNet.
Image Augmentation Using a Task Guided Generative Adversarial Network for Age Estimation on Brain MRI
Aug 03, 2021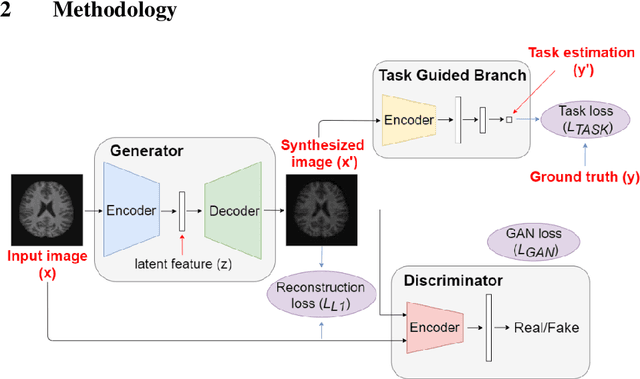
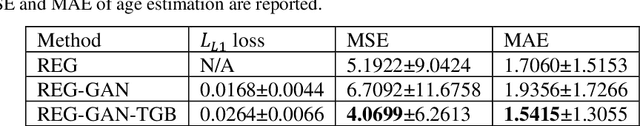

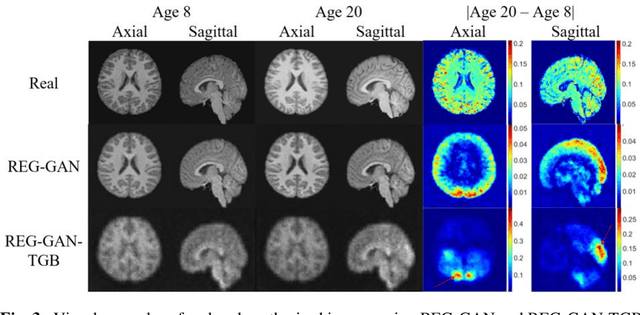
Abstract:Brain age estimation based on magnetic resonance imaging (MRI) is an active research area in early diagnosis of some neurodegenerative diseases (e.g. Alzheimer, Parkinson, Huntington, etc.) for elderly people or brain underdevelopment for the young group. Deep learning methods have achieved the state-of-the-art performance in many medical image analysis tasks, including brain age estimation. However, the performance and generalisability of the deep learning model are highly dependent on the quantity and quality of the training data set. Both collecting and annotating brain MRI data are extremely time-consuming. In this paper, to overcome the data scarcity problem, we propose a generative adversarial network (GAN) based image synthesis method. Different from the existing GAN-based methods, we integrate a task-guided branch (a regression model for age estimation) to the end of the generator in GAN. By adding a task-guided loss to the conventional GAN loss, the learned low-dimensional latent space and the synthesised images are more task-specific. It helps to boost the performance of the down-stream task by combining the synthesised images and real images for model training. The proposed method was evaluated on a public brain MRI data set for age estimation. Our proposed method outperformed (statistically significant) a deep convolutional neural network based regression model and the GAN-based image synthesis method without the task-guided branch. More importantly, it enables the identification of age-related brain regions in the image space. The code is available on GitHub (https://github.com/ruizhe-l/tgb-gan).
FU-net: Multi-class Image Segmentation Using Feedback Weighted U-net
Apr 28, 2020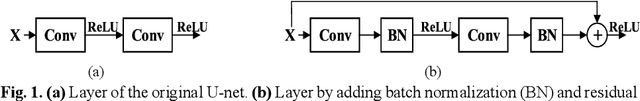
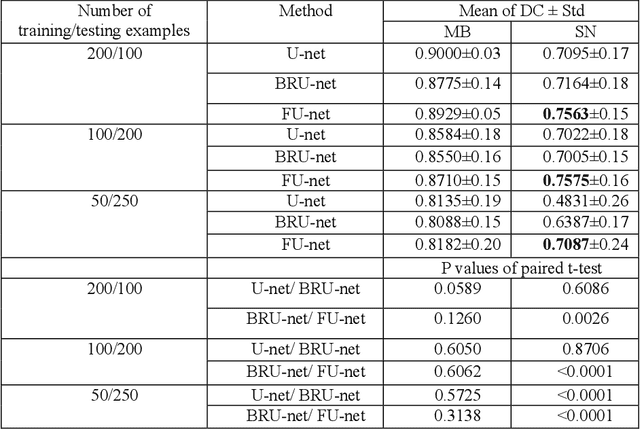

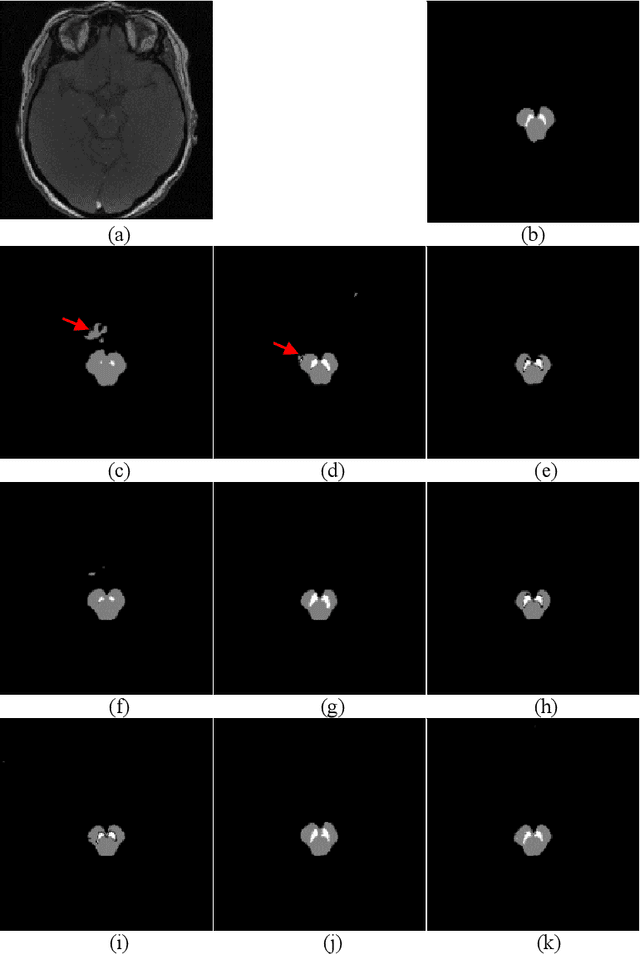
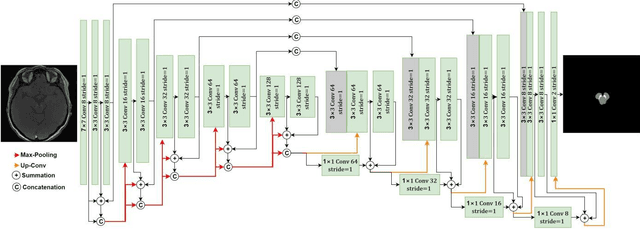
Abstract:In this paper, we present a generic deep convolutional neural network (DCNN) for multi-class image segmentation. It is based on a well-established supervised end-to-end DCNN model, known as U-net. U-net is firstly modified by adding widely used batch normalization and residual block (named as BRU-net) to improve the efficiency of model training. Based on BRU-net, we further introduce a dynamically weighted cross-entropy loss function. The weighting scheme is calculated based on the pixel-wise prediction accuracy during the training process. Assigning higher weights to pixels with lower segmentation accuracies enables the network to learn more from poorly predicted image regions. Our method is named as feedback weighted U-net (FU-net). We have evaluated our method based on T1- weighted brain MRI for the segmentation of midbrain and substantia nigra, where the number of pixels in each class is extremely unbalanced to each other. Based on the dice coefficient measurement, our proposed FU-net has outperformed BRU-net and U-net with statistical significance, especially when only a small number of training examples are available. The code is publicly available in GitHub (GitHub link: https://github.com/MinaJf/FU-net).
* Accepted for publication at International Conference on Image and Graphics (ICIG 2019)
DRU-net: An Efficient Deep Convolutional Neural Network for Medical Image Segmentation
Apr 28, 2020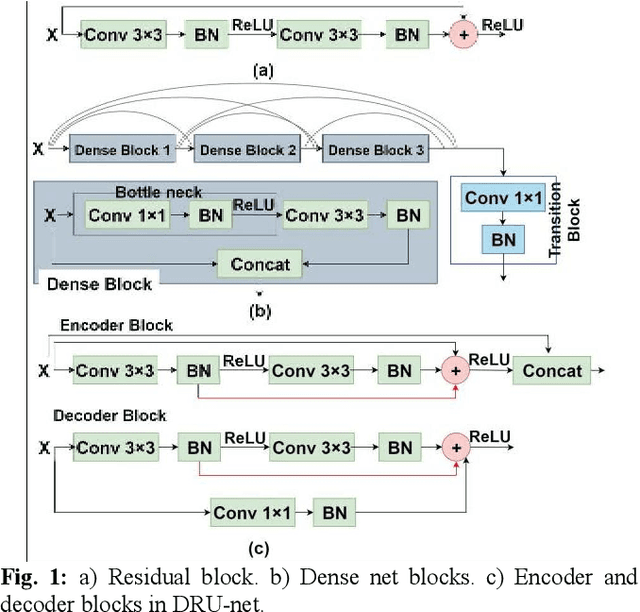
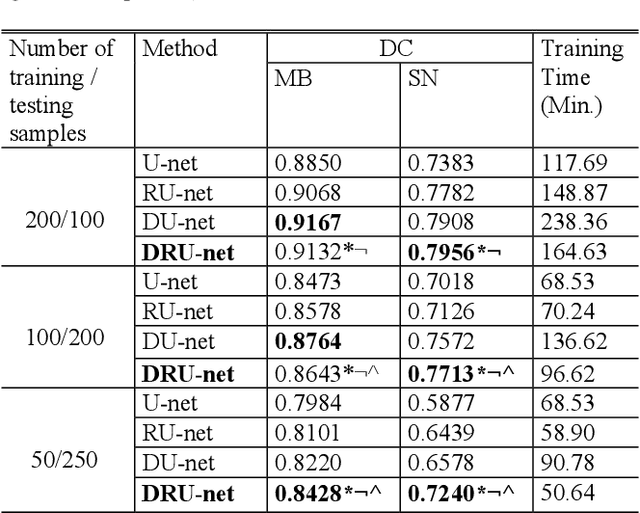
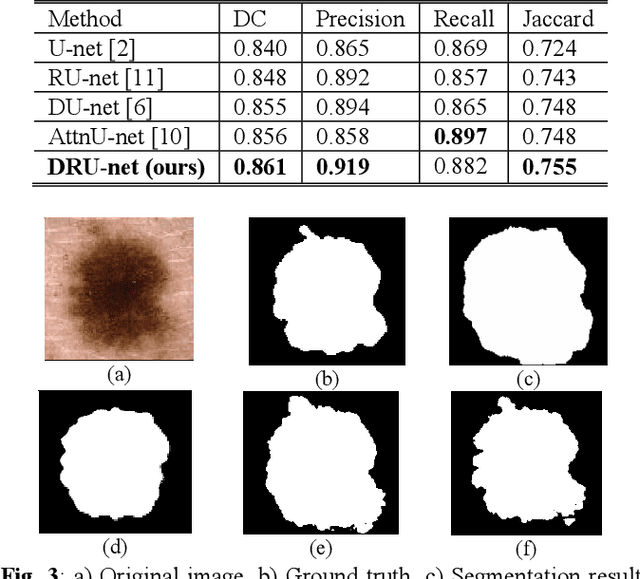
Abstract:Residual network (ResNet) and densely connected network (DenseNet) have significantly improved the training efficiency and performance of deep convolutional neural networks (DCNNs) mainly for object classification tasks. In this paper, we propose an efficient network architecture by considering advantages of both networks. The proposed method is integrated into an encoder-decoder DCNN model for medical image segmentation. Our method adds additional skip connections compared to ResNet but uses significantly fewer model parameters than DenseNet. We evaluate the proposed method on a public dataset (ISIC 2018 grand-challenge) for skin lesion segmentation and a local brain MRI dataset. In comparison with ResNet-based, DenseNet-based and attention network (AttnNet) based methods within the same encoder-decoder network structure, our method achieves significantly higher segmentation accuracy with fewer number of model parameters than DenseNet and AttnNet. The code is available on GitHub (GitHub link: https://github.com/MinaJf/DRU-net).
* Accepted for publication at IEEE International Symposium on Biomedical Imaging (ISBI) 2020, 5 pages, 3 figures
A generic ensemble based deep convolutional neural network for semi-supervised medical image segmentation
Apr 16, 2020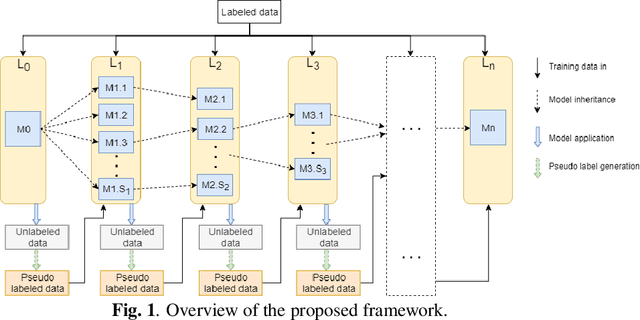
Abstract:Deep learning based image segmentation has achieved the state-of-the-art performance in many medical applications such as lesion quantification, organ detection, etc. However, most of the methods rely on supervised learning, which require a large set of high-quality labeled data. Data annotation is generally an extremely time-consuming process. To address this problem, we propose a generic semi-supervised learning framework for image segmentation based on a deep convolutional neural network (DCNN). An encoder-decoder based DCNN is initially trained using a few annotated training samples. This initially trained model is then copied into sub-models and improved iteratively using random subsets of unlabeled data with pseudo labels generated from models trained in the previous iteration. The number of sub-models is gradually decreased to one in the final iteration. We evaluate the proposed method on a public grand-challenge dataset for skin lesion segmentation. Our method is able to significantly improve beyond fully supervised model learning by incorporating unlabeled data.
* Accepted for publication at IEEE International Symposium on Biomedical Imaging (ISBI) 2020
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge