Fergus V Gleeson
Pinv-Recon: Generalized MR Image Reconstruction via Pseudoinversion of the Encoding Matrix
Oct 08, 2024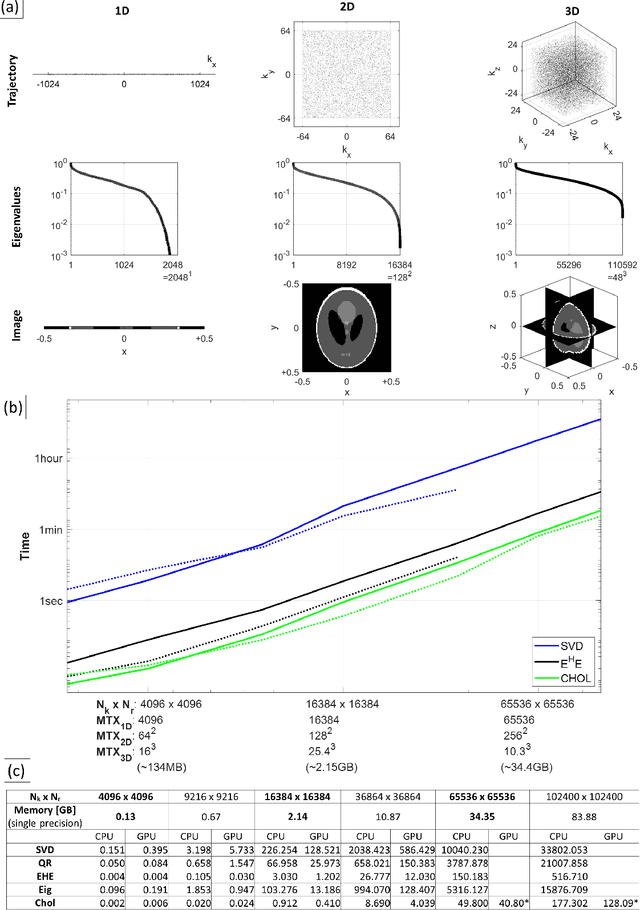
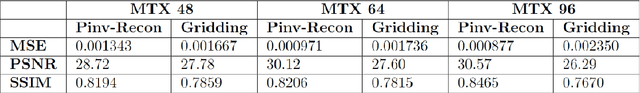
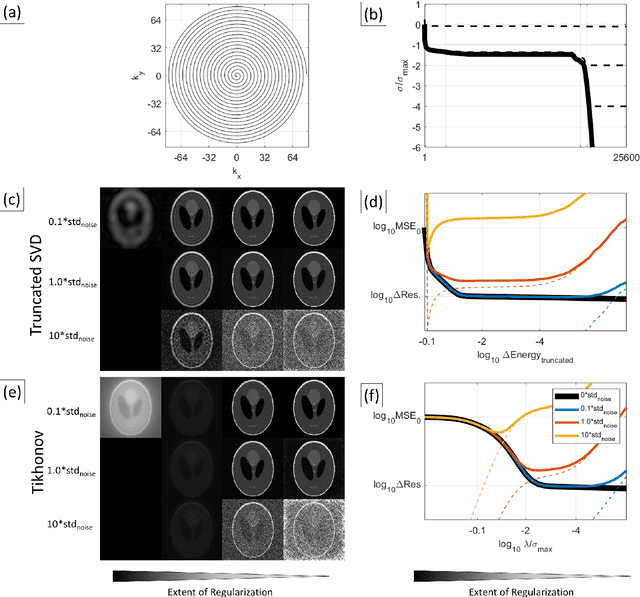
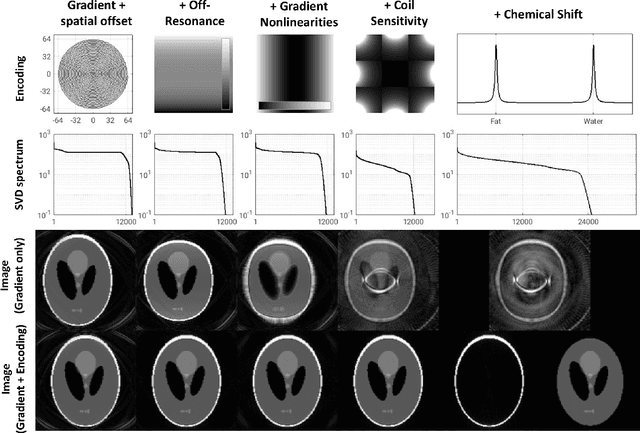
Abstract:Purpose: To present a novel generalized MR image reconstruction based on pseudoinversion of the encoding matrix (Pinv-Recon) as a simple yet powerful method, and demonstrate its computational feasibility for diverse MR imaging applications. Methods: MR image encoding constitutes a linear mapping of the unknown image to the measured k-space data mediated via an encoding matrix ($ data = Encode \times image$). Pinv-Recon addresses MR image reconstruction as a linear inverse problem ($image = Encode^{-1} \times data$), explicitly calculating the Moore-Penrose pseudoinverse of the encoding matrix using truncated singular value decomposition (tSVD). Using a discretized, algebraic notation, we demonstrate constructing a generalized encoding matrix by stacking relevant encoding mechanisms (e.g., gradient encoding, coil sensitivity encoding, chemical shift inversion) and encoding distortions (e.g., off-center positioning, B$_0$ inhomogeneity, spatiotemporal gradient imperfections, transient relaxation effects). Iterative reconstructions using the explicit generalized encoding matrix, and the computation of the spatial-response-function (SRF) and noise amplification, were demonstrated. Results: We evaluated the computation times and memory requirements (time ~ (size of the encoding matrix)$^{1.4}$). Using the Shepp-Logan phantom, we demonstrated the versatility of the method for various intertwined MR image encoding and distortion mechanisms, achieving better MSE, PSNR and SSIM metrics than conventional methods. A diversity of datasets, including the ISMRM CG-SENSE challenge, were used to validate Pinv-Recon. Conclusion: Although pseudo-inversion of large encoding matrices was once deemed computationally intractable, recent advances make Pinv-Recon feasible. It has great promise for both research and clinical applications, and for educational use.
Pieces-of-parts for supervoxel segmentation with global context: Application to DCE-MRI tumour delineation
Apr 18, 2016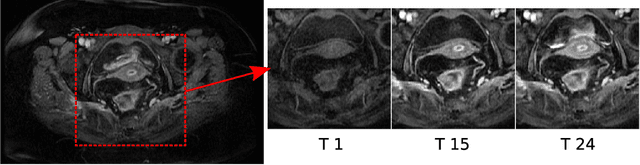

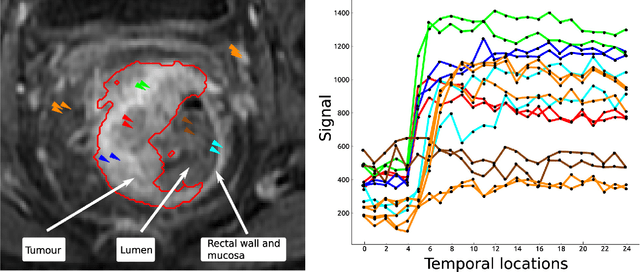
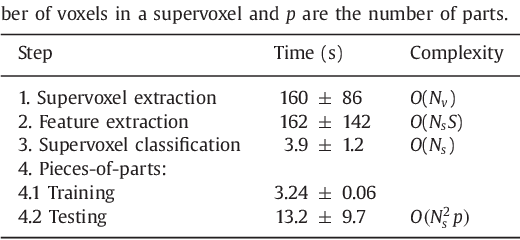
Abstract:Rectal tumour segmentation in dynamic contrast-enhanced MRI (DCE-MRI) is a challenging task, and an automated and consistent method would be highly desirable to improve the modelling and prediction of patient outcomes from tissue contrast enhancement characteristics - particularly in routine clinical practice. A framework is developed to automate DCE-MRI tumour segmentation, by introducing: perfusion-supervoxels to over-segment and classify DCE-MRI volumes using the dynamic contrast enhancement characteristics; and the pieces-of-parts graphical model, which adds global (anatomic) constraints that further refine the supervoxel components that comprise the tumour. The framework was evaluated on 23 DCE-MRI scans of patients with rectal adenocarcinomas, and achieved a voxelwise area-under the receiver operating characteristic curve (AUC) of 0.97 compared to expert delineations. Creating a binary tumour segmentation, 21 of the 23 cases were segmented correctly with a median Dice similarity coefficient (DSC) of 0.63, which is close to the inter-rater variability of this challenging task. A sec- ond study is also included to demonstrate the method's generalisability and achieved a DSC of 0.71. The framework achieves promising results for the underexplored area of rectal tumour segmentation in DCE-MRI, and the methods have potential to be applied to other DCE-MRI and supervoxel segmentation problems
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge