Donniell E. Fishkind
Seeded Graph Matching
Apr 10, 2018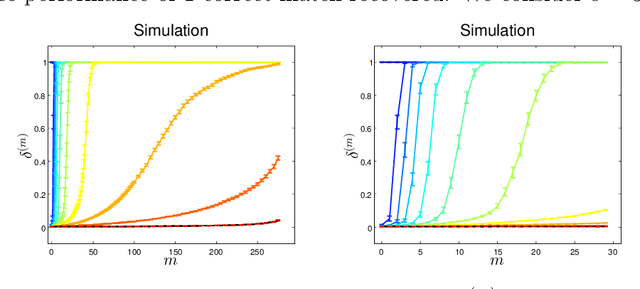
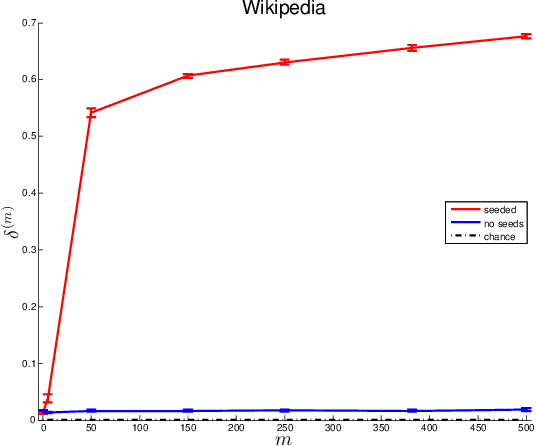

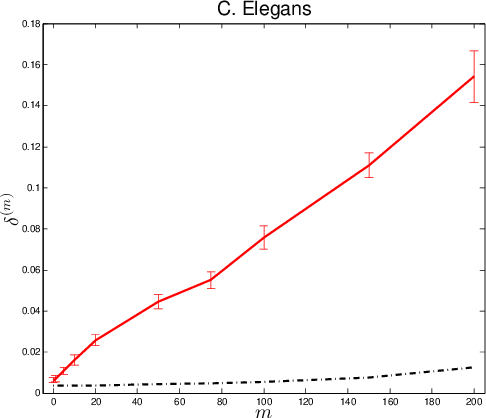
Abstract:Given two graphs, the graph matching problem is to align the two vertex sets so as to minimize the number of adjacency disagreements between the two graphs. The seeded graph matching problem is the graph matching problem when we are first given a partial alignment that we are tasked with completing. In this paper, we modify the state-of-the-art approximate graph matching algorithm "FAQ" of Vogelstein et al. (2015) to make it a fast approximate seeded graph matching algorithm, adapt its applicability to include graphs with differently sized vertex sets, and extend the algorithm so as to provide, for each individual vertex, a nomination list of likely matches. We demonstrate the effectiveness of our algorithm via simulation and real data experiments; indeed, knowledge of even a few seeds can be extremely effective when our seeded graph matching algorithm is used to recover a naturally existing alignment that is only partially observed.
Statistical inference on random dot product graphs: a survey
Sep 16, 2017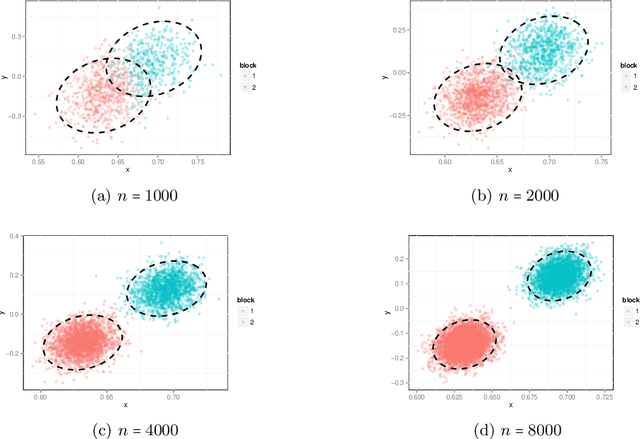
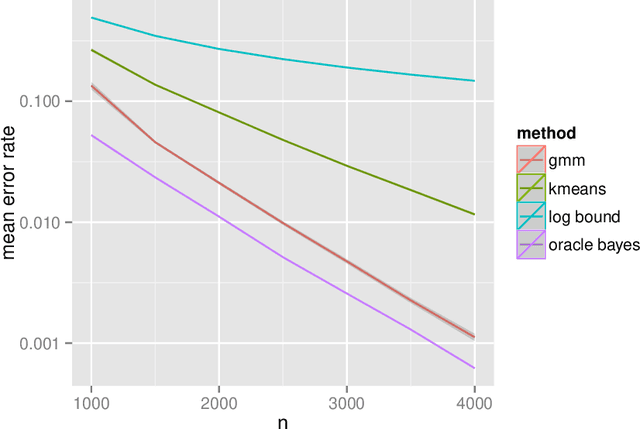

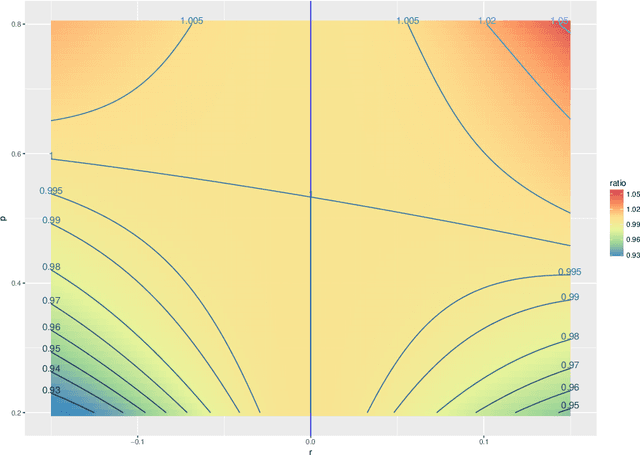
Abstract:The random dot product graph (RDPG) is an independent-edge random graph that is analytically tractable and, simultaneously, either encompasses or can successfully approximate a wide range of random graphs, from relatively simple stochastic block models to complex latent position graphs. In this survey paper, we describe a comprehensive paradigm for statistical inference on random dot product graphs, a paradigm centered on spectral embeddings of adjacency and Laplacian matrices. We examine the analogues, in graph inference, of several canonical tenets of classical Euclidean inference: in particular, we summarize a body of existing results on the consistency and asymptotic normality of the adjacency and Laplacian spectral embeddings, and the role these spectral embeddings can play in the construction of single- and multi-sample hypothesis tests for graph data. We investigate several real-world applications, including community detection and classification in large social networks and the determination of functional and biologically relevant network properties from an exploratory data analysis of the Drosophila connectome. We outline requisite background and current open problems in spectral graph inference.
* An expository survey paper on a comprehensive paradigm for inference for random dot product graphs, centered on graph adjacency and Laplacian spectral embeddings. Paper outlines requisite background; summarizes theory, methodology, and applications from previous and ongoing work; and closes with a discussion of several open problems
On the Consistency of the Likelihood Maximization Vertex Nomination Scheme: Bridging the Gap Between Maximum Likelihood Estimation and Graph Matching
Aug 27, 2016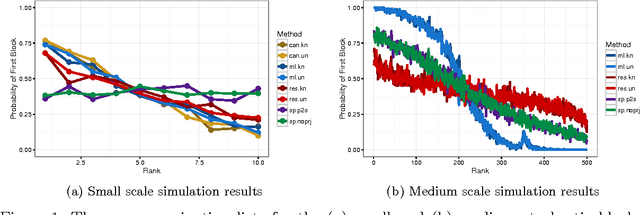

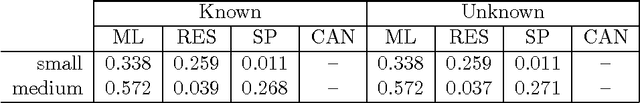

Abstract:Given a graph in which a few vertices are deemed interesting a priori, the vertex nomination task is to order the remaining vertices into a nomination list such that there is a concentration of interesting vertices at the top of the list. Previous work has yielded several approaches to this problem, with theoretical results in the setting where the graph is drawn from a stochastic block model (SBM), including a vertex nomination analogue of the Bayes optimal classifier. In this paper, we prove that maximum likelihood (ML)-based vertex nomination is consistent, in the sense that the performance of the ML-based scheme asymptotically matches that of the Bayes optimal scheme. We prove theorems of this form both when model parameters are known and unknown. Additionally, we introduce and prove consistency of a related, more scalable restricted-focus ML vertex nomination scheme. Finally, we incorporate vertex and edge features into ML-based vertex nomination and briefly explore the empirical effectiveness of this approach.
Spectral Clustering for Divide-and-Conquer Graph Matching
Mar 12, 2015Abstract:We present a parallelized bijective graph matching algorithm that leverages seeds and is designed to match very large graphs. Our algorithm combines spectral graph embedding with existing state-of-the-art seeded graph matching procedures. We justify our approach by proving that modestly correlated, large stochastic block model random graphs are correctly matched utilizing very few seeds through our divide-and-conquer procedure. We also demonstrate the effectiveness of our approach in matching very large graphs in simulated and real data examples, showing up to a factor of 8 improvement in runtime with minimal sacrifice in accuracy.
On the Incommensurability Phenomenon
Feb 06, 2015


Abstract:Suppose that two large, multi-dimensional data sets are each noisy measurements of the same underlying random process, and principle components analysis is performed separately on the data sets to reduce their dimensionality. In some circumstances it may happen that the two lower-dimensional data sets have an inordinately large Procrustean fitting-error between them. The purpose of this manuscript is to quantify this "incommensurability phenomenon." In particular, under specified conditions, the square Procrustean fitting-error of the two normalized lower-dimensional data sets is (asymptotically) a convex combination (via a correlation parameter) of the Hausdorff distance between the projection subspaces and the maximum possible value of the square Procrustean fitting-error for normalized data. We show how this gives rise to the incommensurability phenomenon, and we employ illustrative simulations as well as a real data experiment to explore how the incommensurability phenomenon may have an appreciable impact.
A consistent adjacency spectral embedding for stochastic blockmodel graphs
Apr 27, 2012



Abstract:We present a method to estimate block membership of nodes in a random graph generated by a stochastic blockmodel. We use an embedding procedure motivated by the random dot product graph model, a particular example of the latent position model. The embedding associates each node with a vector; these vectors are clustered via minimization of a square error criterion. We prove that this method is consistent for assigning nodes to blocks, as only a negligible number of nodes will be mis-assigned. We prove consistency of the method for directed and undirected graphs. The consistent block assignment makes possible consistent parameter estimation for a stochastic blockmodel. We extend the result in the setting where the number of blocks grows slowly with the number of nodes. Our method is also computationally feasible even for very large graphs. We compare our method to Laplacian spectral clustering through analysis of simulated data and a graph derived from Wikipedia documents.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge