Donal Landers
SemEval-2023 Task 7: Multi-Evidence Natural Language Inference for Clinical Trial Data
May 11, 2023Abstract:This paper describes the results of SemEval 2023 task 7 -- Multi-Evidence Natural Language Inference for Clinical Trial Data (NLI4CT) -- consisting of 2 tasks, a Natural Language Inference (NLI) task, and an evidence selection task on clinical trial data. The proposed challenges require multi-hop biomedical and numerical reasoning, which are of significant importance to the development of systems capable of large-scale interpretation and retrieval of medical evidence, to provide personalized evidence-based care. Task 1, the entailment task, received 643 submissions from 40 participants, and Task 2, the evidence selection task, received 364 submissions from 23 participants. The tasks are challenging, with the majority of submitted systems failing to significantly outperform the majority class baseline on the entailment task, and we observe significantly better performance on the evidence selection task than on the entailment task. Increasing the number of model parameters leads to a direct increase in performance, far more significant than the effect of biomedical pre-training. Future works could explore the limitations of large models for generalization and numerical inference, and investigate methods to augment clinical datasets to allow for more rigorous testing and to facilitate fine-tuning. We envisage that the dataset, models, and results of this task will be useful to the biomedical NLI and evidence retrieval communities. The dataset, competition leaderboard, and website are publicly available.
NLI4CT: Multi-Evidence Natural Language Inference for Clinical Trial Reports
May 05, 2023



Abstract:How can we interpret and retrieve medical evidence to support clinical decisions? Clinical trial reports (CTR) amassed over the years contain indispensable information for the development of personalized medicine. However, it is practically infeasible to manually inspect over 400,000+ clinical trial reports in order to find the best evidence for experimental treatments. Natural Language Inference (NLI) offers a potential solution to this problem, by allowing the scalable computation of textual entailment. However, existing NLI models perform poorly on biomedical corpora, and previously published datasets fail to capture the full complexity of inference over CTRs. In this work, we present a novel resource to advance research on NLI for reasoning on CTRs. The resource includes two main tasks. Firstly, to determine the inference relation between a natural language statement, and a CTR. Secondly, to retrieve supporting facts to justify the predicted relation. We provide NLI4CT, a corpus of 2400 statements and CTRs, annotated for these tasks. Baselines on this corpus expose the limitations of existing NLI models, with 6 state-of-the-art NLI models achieving a maximum F1 score of 0.627. To the best of our knowledge, we are the first to design a task that covers the interpretation of full CTRs. To encourage further work on this challenging dataset, we make the corpus, competition leaderboard, website and code to replicate the baseline experiments available at: https://github.com/ai-systems/nli4ct
Metareview-informed Explainable Cytokine Storm Detection during CAR-T cell Therapy
Jun 20, 2022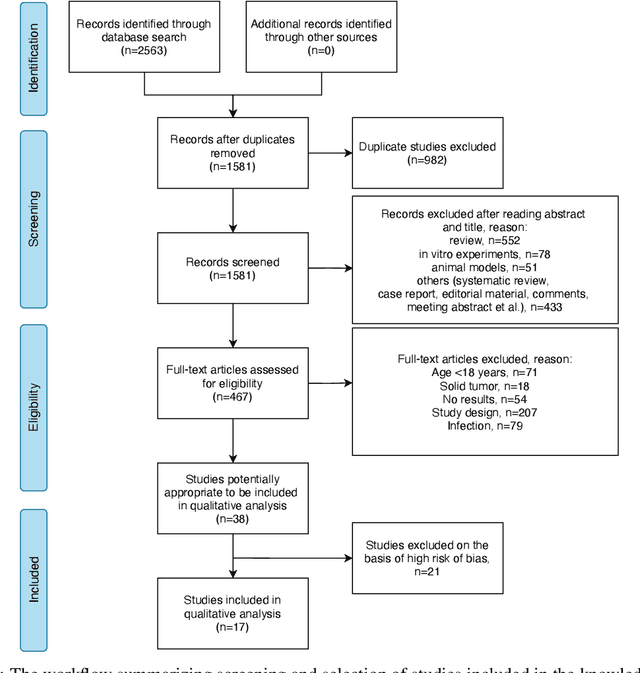
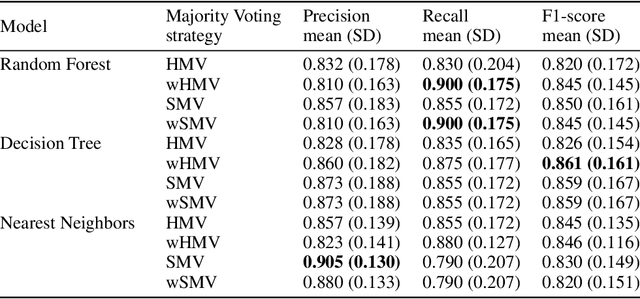
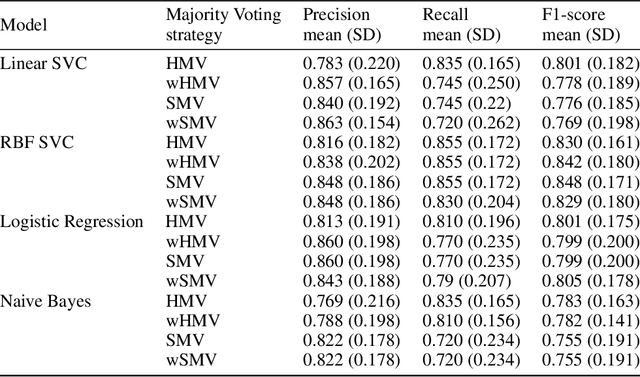
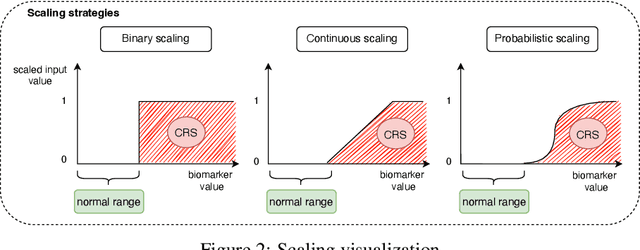
Abstract:Cytokine release syndrome (CRS), also known as cytokine storm, is one of the most consequential adverse effects of chimeric antigen receptor therapies that have shown promising results in cancer treatment. When emerging, CRS could be identified by the analysis of specific cytokine and chemokine profiles that tend to exhibit similarities across patients. In this paper, we exploit these similarities using machine learning algorithms and set out to pioneer a meta--review informed method for the identification of CRS based on specific cytokine peak concentrations and evidence from previous clinical studies. We argue that such methods could support clinicians in analyzing suspect cytokine profiles by matching them against CRS knowledge from past clinical studies, with the ultimate aim of swift CRS diagnosis. During evaluation with real--world CRS clinical data, we emphasize the potential of our proposed method of producing interpretable results, in addition to being effective in identifying the onset of cytokine storm.
Architectures of Meaning, A Systematic Corpus Analysis of NLP Systems
Jul 16, 2021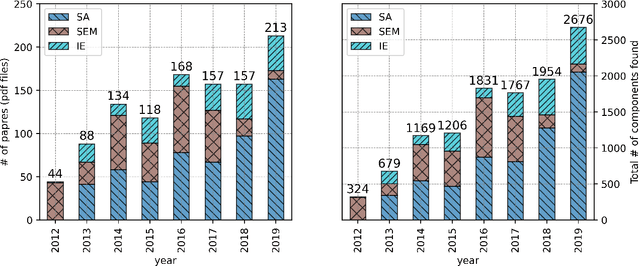
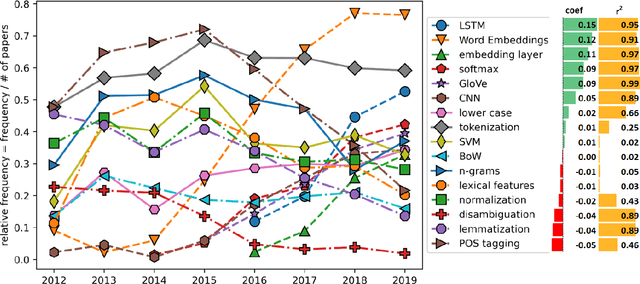
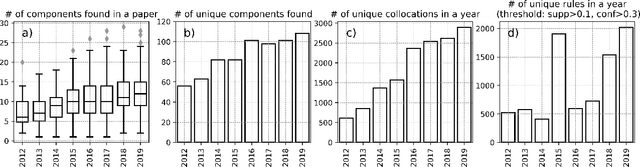
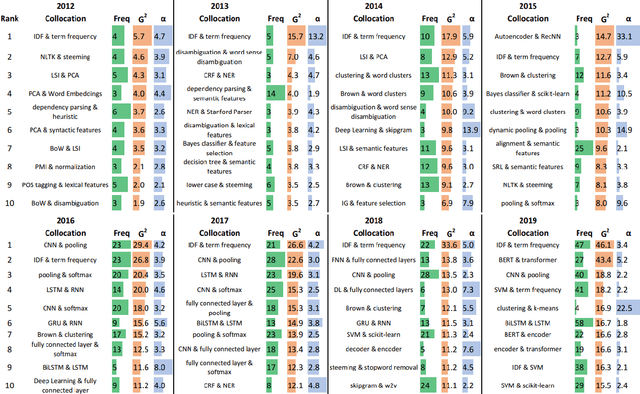
Abstract:This paper proposes a novel statistical corpus analysis framework targeted towards the interpretation of Natural Language Processing (NLP) architectural patterns at scale. The proposed approach combines saturation-based lexicon construction, statistical corpus analysis methods and graph collocations to induce a synthesis representation of NLP architectural patterns from corpora. The framework is validated in the full corpus of Semeval tasks and demonstrated coherent architectural patterns which can be used to answer architectural questions on a data-driven fashion, providing a systematic mechanism to interpret a largely dynamic and exponentially growing field.
Encoding Explanatory Knowledge for Zero-shot Science Question Answering
May 19, 2021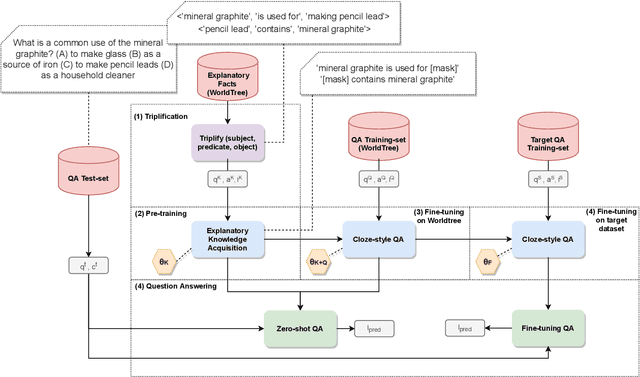
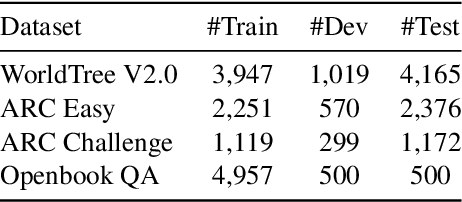
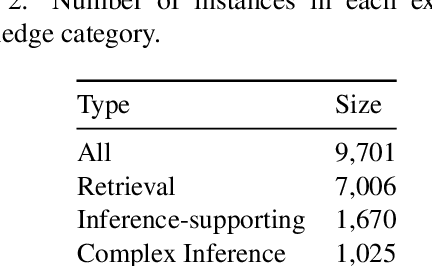
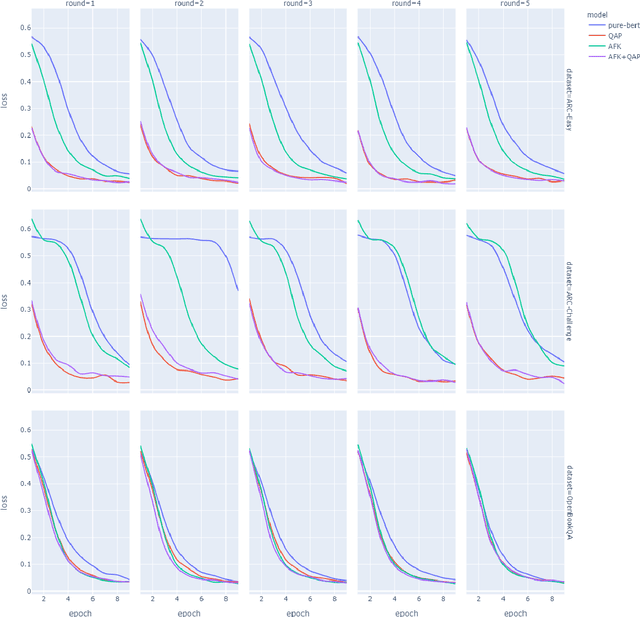
Abstract:This paper describes N-XKT (Neural encoding based on eXplanatory Knowledge Transfer), a novel method for the automatic transfer of explanatory knowledge through neural encoding mechanisms. We demonstrate that N-XKT is able to improve accuracy and generalization on science Question Answering (QA). Specifically, by leveraging facts from background explanatory knowledge corpora, the N-XKT model shows a clear improvement on zero-shot QA. Furthermore, we show that N-XKT can be fine-tuned on a target QA dataset, enabling faster convergence and more accurate results. A systematic analysis is conducted to quantitatively analyze the performance of the N-XKT model and the impact of different categories of knowledge on the zero-shot generalization task.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge