Dillon Laird
MURA: Large Dataset for Abnormality Detection in Musculoskeletal Radiographs
May 22, 2018
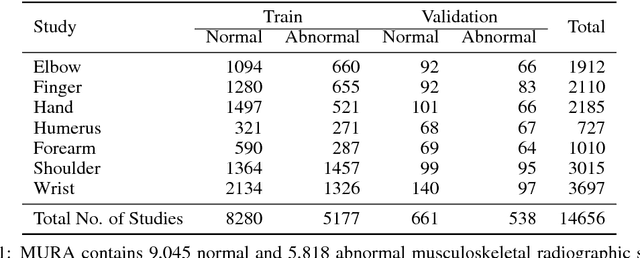
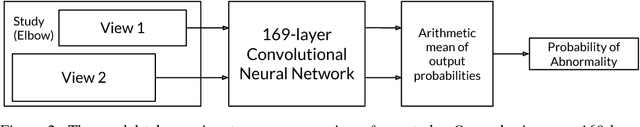
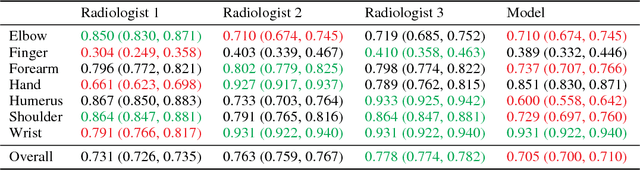
Abstract:We introduce MURA, a large dataset of musculoskeletal radiographs containing 40,561 images from 14,863 studies, where each study is manually labeled by radiologists as either normal or abnormal. To evaluate models robustly and to get an estimate of radiologist performance, we collect additional labels from six board-certified Stanford radiologists on the test set, consisting of 207 musculoskeletal studies. On this test set, the majority vote of a group of three radiologists serves as gold standard. We train a 169-layer DenseNet baseline model to detect and localize abnormalities. Our model achieves an AUROC of 0.929, with an operating point of 0.815 sensitivity and 0.887 specificity. We compare our model and radiologists on the Cohen's kappa statistic, which expresses the agreement of our model and of each radiologist with the gold standard. Model performance is comparable to the best radiologist performance in detecting abnormalities on finger and wrist studies. However, model performance is lower than best radiologist performance in detecting abnormalities on elbow, forearm, hand, humerus, and shoulder studies. We believe that the task is a good challenge for future research. To encourage advances, we have made our dataset freely available at https://stanfordmlgroup.github.io/competitions/mura .
Stochastic Variational Inference for Hidden Markov Models
Nov 06, 2014

Abstract:Variational inference algorithms have proven successful for Bayesian analysis in large data settings, with recent advances using stochastic variational inference (SVI). However, such methods have largely been studied in independent or exchangeable data settings. We develop an SVI algorithm to learn the parameters of hidden Markov models (HMMs) in a time-dependent data setting. The challenge in applying stochastic optimization in this setting arises from dependencies in the chain, which must be broken to consider minibatches of observations. We propose an algorithm that harnesses the memory decay of the chain to adaptively bound errors arising from edge effects. We demonstrate the effectiveness of our algorithm on synthetic experiments and a large genomics dataset where a batch algorithm is computationally infeasible.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge