Davide Poggiali
Interpolation with the polynomial kernels
Dec 15, 2022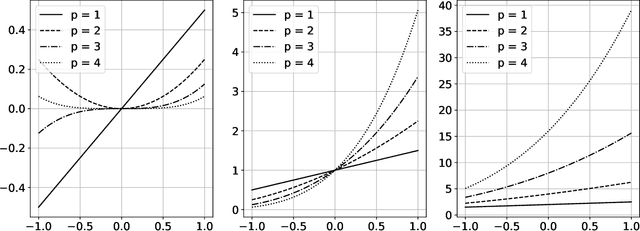
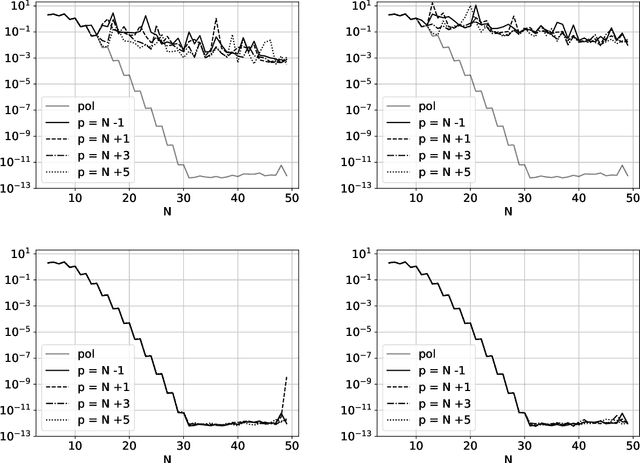
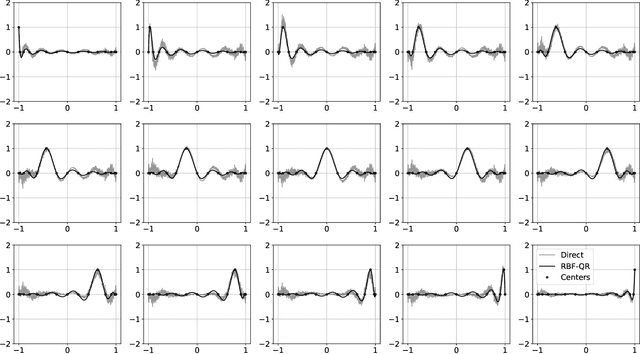
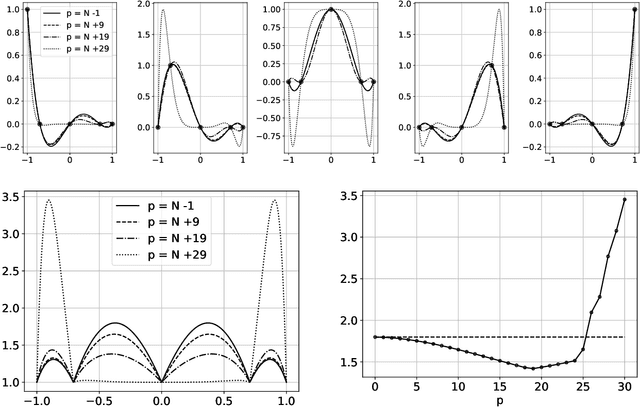
Abstract:The polynomial kernels are widely used in machine learning and they are one of the default choices to develop kernel-based classification and regression models. However, they are rarely used and considered in numerical analysis due to their lack of strict positive definiteness. In particular they do not enjoy the usual property of unisolvency for arbitrary point sets, which is one of the key properties used to build kernel-based interpolation methods. This paper is devoted to establish some initial results for the study of these kernels, and their related interpolation algorithms, in the context of approximation theory. We will first prove necessary and sufficient conditions on point sets which guarantee the existence and uniqueness of an interpolant. We will then study the Reproducing Kernel Hilbert Spaces (or native spaces) of these kernels and their norms, and provide inclusion relations between spaces corresponding to different kernel parameters. With these spaces at hand, it will be further possible to derive generic error estimates which apply to sufficiently smooth functions, thus escaping the native space. Finally, we will show how to employ an efficient stable algorithm to these kernels to obtain accurate interpolants, and we will test them in some numerical experiment. After this analysis several computational and theoretical aspects remain open, and we will outline possible further research directions in a concluding section. This work builds some bridges between kernel and polynomial interpolation, two topics to which the authors, to different extents, have been introduced under the supervision or through the work of Stefano De Marchi. For this reason, they wish to dedicate this work to him in the occasion of his 60th birthday.
Reducing the Gibbs effect in multimodal medical imaging by the Fake Nodes Approach
Feb 21, 2022



Abstract:It is a common practice in multimodal medical imaging to undersample the anatomically-derived segmentation images to measure the mean activity of a co-acquired functional image. This practice avoids the resampling-related Gibbs effect that would occur in oversampling the functional image. As sides effect, waste of time and efforts are produced since the anatomical segmentation at full resolution is performed in many hours of computations or manual work. In this work we explain the commonly-used resampling methods and give errors bound in the cases of continuous and discontinuous signals. Then we propose a Fake Nodes scheme for image resampling designed to reduce the Gibbs effect when oversampling the functional image. This new approach is compared to the traditional counterpart in two significant experiments, both showing that Fake Nodes resampling gives smaller errors.
Oversampling errors in multimodal medical imaging are due to the Gibbs effect
Mar 10, 2021



Abstract:To analyse multimodal 3-dimensional medical images, interpolation is required for resampling which - unavoidably - introduces an interpolation error. In this work we consider three segmented 3-dimensional images resampled with three different neuroimaging software tools for comparing undersampling and oversampling strategies and to identify where the oversampling error lies. The results indicate that undersampling to the lowest image size is advantageous in terms of mean value per segment errors and that the oversampling error is larger where the gradient is steeper, showing a Gibbs effect.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge