David B. Grayden
Decoding Saccadic Eye Movements from Brain Signals Using an Endovascular Neural Interface
Jun 09, 2025Abstract:An Oculomotor Brain-Computer Interface (BCI) records neural activity from regions of the brain involved in planning eye movements and translates this activity into control commands. While previous successful oculomotor BCI studies primarily relied on invasive microelectrode implants in non-human primates, this study investigates the feasibility of an oculomotor BCI using a minimally invasive endovascular Stentrode device implanted near the supplementary motor area in a patient with amyotrophic lateral sclerosis (ALS). To achieve this, self-paced visually-guided and free-viewing saccade tasks were designed, in which the participant performed saccades in four directions (left, right, up, down), with simultaneous recording of endovascular EEG and eye gaze. The visually guided saccades were cued with visual stimuli, whereas the free-viewing saccades were self-directed without explicit cues. The results showed that while the neural responses of visually guided saccades overlapped with the cue-evoked potentials, the free-viewing saccades exhibited distinct saccade-related potentials that began shortly before eye movement, peaked approximately 50 ms after saccade onset, and persisted for around 200 ms. In the frequency domain, these responses appeared as a low-frequency synchronisation below 15 Hz. Classification of 'fixation vs. saccade' was robust, achieving mean area under the receiver operating characteristic curve (AUC) scores of 0.88 within sessions and 0.86 between sessions. In contrast, classifying saccade direction proved more challenging, yielding within-session AUC scores of 0.67 for four-class decoding and up to 0.75 for the best-performing binary comparisons (left vs. up and left vs. down). This proof-of-concept study demonstrates the feasibility of an endovascular oculomotor BCI in an ALS patient, establishing a foundation for future oculomotor BCI studies in human subjects.
Sub-Scalp Brain-Computer Interface Device Design and Fabrication
Apr 17, 2025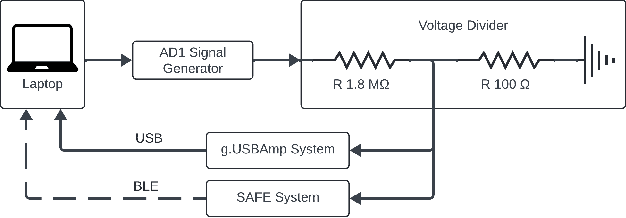
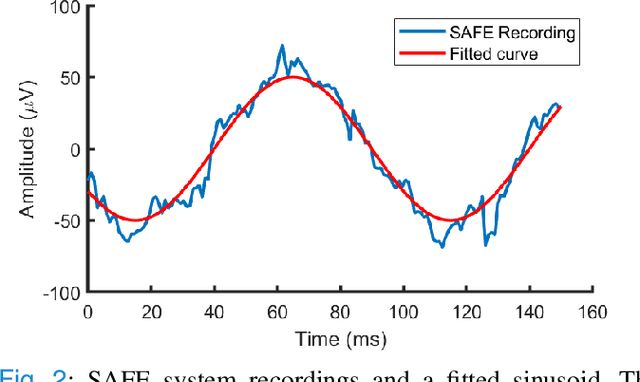
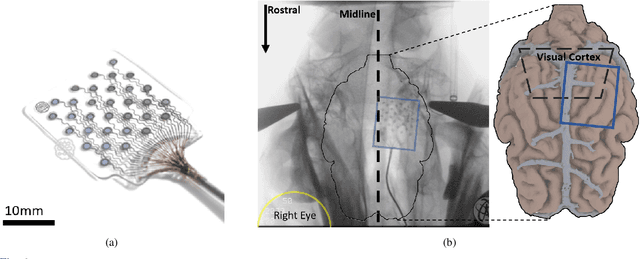
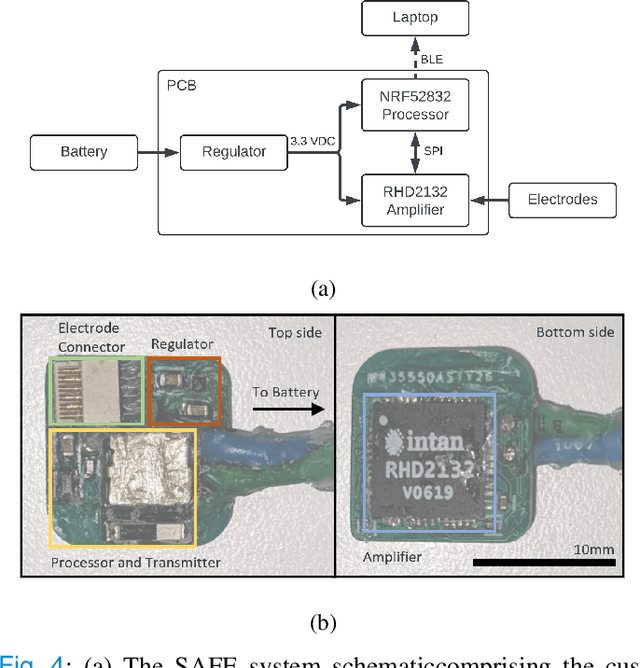
Abstract:Current brain-computer interfaces (BCI) face limitations in signal acquisition. While sub-scalp EEG offers a potential solution, existing devices prioritize chronic seizure monitoring and lack features suited for BCI applications. This work addresses this gap by outlining key specifications for sub-scalp BCI devices, focusing on channel count, sampling rate, power efficiency, and form factor. We present the Set-And-Forget EEG (SAFE) system, a custom-built amplifier and wireless transmitter meeting these criteria. This compact (12x12 mm), six-channel device offers 1024 Hz sampling and Bluetooth Low Energy data transmission. Validation using generated sinusoids and electrocorticography recordings of visual evoked potentials in sheep models demonstrated low noise recording. Future animal studies will assess sub-scalp EEG signal quality for BCI applications. This data lays the groundwork for human trials, ultimately paving the way for chronic, in-home BCIs that empower individuals with physical disabilities.
Path Signatures for Seizure Forecasting
Aug 18, 2023Abstract:Forecasting the state of a system from an observed time series is the subject of research in many domains, such as computational neuroscience. Here, the prediction of epileptic seizures from brain measurements is an unresolved problem. There are neither complete models describing underlying brain dynamics, nor do individual patients exhibit a single seizure onset pattern, which complicates the development of a `one-size-fits-all' solution. Based on a longitudinal patient data set, we address the automated discovery and quantification of statistical features (biomarkers) that can be used to forecast seizures in a patient-specific way. We use existing and novel feature extraction algorithms, in particular the path signature, a recent development in time series analysis. Of particular interest is how this set of complex, nonlinear features performs compared to simpler, linear features on this task. Our inference is based on statistical classification algorithms with in-built subset selection to discern time series with and without an impending seizure while selecting only a small number of relevant features. This study may be seen as a step towards a generalisable pattern recognition pipeline for time series in a broader context.
Autoregressive models for biomedical signal processing
May 01, 2023Abstract:Autoregressive models are ubiquitous tools for the analysis of time series in many domains such as computational neuroscience and biomedical engineering. In these domains, data is, for example, collected from measurements of brain activity. Crucially, this data is subject to measurement errors as well as uncertainties in the underlying system model. As a result, standard signal processing using autoregressive model estimators may be biased. We present a framework for autoregressive modelling that incorporates these uncertainties explicitly via an overparameterised loss function. To optimise this loss, we derive an algorithm that alternates between state and parameter estimation. Our work shows that the procedure is able to successfully denoise time series and successfully reconstruct system parameters. This new paradigm can be used in a multitude of applications in neuroscience such as brain-computer interface data analysis and better understanding of brain dynamics in diseases such as epilepsy.
Brain Model State Space Reconstruction Using an LSTM Neural Network
Jan 20, 2023Abstract:Objective Kalman filtering has previously been applied to track neural model states and parameters, particularly at the scale relevant to EEG. However, this approach lacks a reliable method to determine the initial filter conditions and assumes that the distribution of states remains Gaussian. This study presents an alternative, data-driven method to track the states and parameters of neural mass models (NMMs) from EEG recordings using deep learning techniques, specifically an LSTM neural network. Approach An LSTM filter was trained on simulated EEG data generated by a neural mass model using a wide range of parameters. With an appropriately customised loss function, the LSTM filter can learn the behaviour of NMMs. As a result, it can output the state vector and parameters of NMMs given observation data as the input. Main Results Test results using simulated data yielded correlations with R squared of around 0.99 and verified that the method is robust to noise and can be more accurate than a nonlinear Kalman filter when the initial conditions of the Kalman filter are not accurate. As an example of real-world application, the LSTM filter was also applied to real EEG data that included epileptic seizures, and revealed changes in connectivity strength parameters at the beginnings of seizures. Significance Tracking the state vector and parameters of mathematical brain models is of great importance in the area of brain modelling, monitoring, imaging and control. This approach has no need to specify the initial state vector and parameters, which is very difficult to do in practice because many of the variables being estimated cannot be measured directly in physiological experiments. This method may be applied using any neural mass model and, therefore, provides a general, novel, efficient approach to estimate brain model variables that are often difficult to measure.
Frequency Superposition -- A Multi-Frequency Stimulation Method in SSVEP-based BCIs
Apr 25, 2021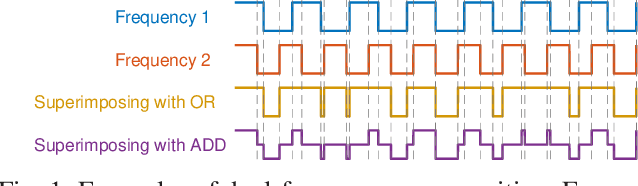
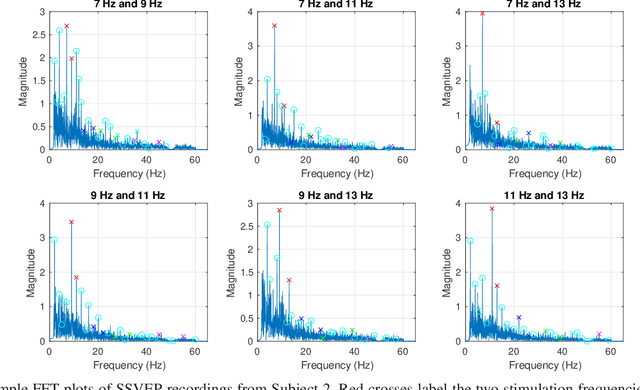
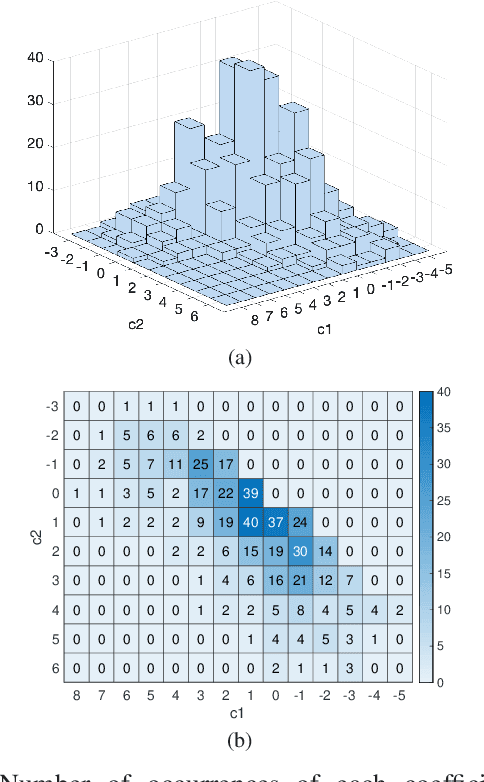
Abstract:The steady-state visual evoked potential (SSVEP) is one of the most widely used modalities in brain-computer interfaces (BCIs) due to its many advantages. However, the existence of harmonics and the limited range of responsive frequencies in SSVEP make it challenging to further expand the number of targets without sacrificing other aspects of the interface or putting additional constraints on the system. This paper introduces a novel multi-frequency stimulation method for SSVEP and investigates its potential to effectively and efficiently increase the number of targets presented. The proposed stimulation method, obtained by the superposition of the stimulation signals at different frequencies, is size-efficient, allows single-step target identification, puts no strict constraints on the usable frequency range, can be suited to self-paced BCIs, and does not require specific light sources. In addition to the stimulus frequencies and their harmonics, the evoked SSVEP waveforms include frequencies that are integer linear combinations of the stimulus frequencies. Results of decoding SSVEPs collected from nine subjects using canonical correlation analysis (CCA) with only the frequencies and harmonics as reference, also demonstrate the potential of using such a stimulation paradigm in SSVEP-based BCIs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge