Danilo Silva
Towards a more realistic evaluation of machine learning models for bearing fault diagnosis
Sep 26, 2025



Abstract:Reliable detection of bearing faults is essential for maintaining the safety and operational efficiency of rotating machinery. While recent advances in machine learning (ML), particularly deep learning, have shown strong performance in controlled settings, many studies fail to generalize to real-world applications due to methodological flaws, most notably data leakage. This paper investigates the issue of data leakage in vibration-based bearing fault diagnosis and its impact on model evaluation. We demonstrate that common dataset partitioning strategies, such as segment-wise and condition-wise splits, introduce spurious correlations that inflate performance metrics. To address this, we propose a rigorous, leakage-free evaluation methodology centered on bearing-wise data partitioning, ensuring no overlap between the physical components used for training and testing. Additionally, we reformulate the classification task as a multi-label problem, enabling the detection of co-occurring fault types and the use of prevalence-independent metrics such as Macro AUROC. Beyond preventing leakage, we also examine the effect of dataset diversity on generalization, showing that the number of unique training bearings is a decisive factor for achieving robust performance. We evaluate our methodology on three widely adopted datasets: CWRU, Paderborn University (PU), and University of Ottawa (UORED-VAFCLS). This study highlights the importance of leakage-aware evaluation protocols and provides practical guidelines for dataset partitioning, model selection, and validation, fostering the development of more trustworthy ML systems for industrial fault diagnosis applications.
Benchmarking Deep Learning-Based Methods for Irradiance Nowcasting with Sky Images
Mar 27, 2025



Abstract:To address the high levels of uncertainty associated with photovoltaic energy, an increasing number of studies focusing on short-term solar forecasting have been published. Most of these studies use deep learning-based models to directly forecast a solar irradiance or photovoltaic power value given an input of sky image sequences. Recently, however, advances in generative modeling have led to approaches that divide the forecasting problem into two sub-problems: 1) future event prediction, i.e. generating future sky images; and 2) solar irradiance or photovoltaic power nowcasting, i.e. predicting the concurrent value from a single image. One such approach is the SkyGPT model, where they show that the potential for improvement is much larger for the nowcasting model than for the generative model. Thus, in this paper, we focus on the solar irradiance nowcasting problem and conduct an extensive benchmark of deep learning architectures across the widely-used Folsom, SIRTA and NREL datasets. Moreover, we perform ablation experiments on different training configurations and data processing techniques, including the choice of the target variable used for training and adjustments of the timestamp alignment between images and irradiance measurements. In particular, we draw attention to a potential error associated with the sky image timestamps in the Folsom dataset and a possible fix is discussed. All our results are reported in terms of both the root mean squared error and the mean absolute error and, by leveraging the three datasets, we demonstrate that our findings are consistent across different solar stations.
Benchmarking deep learning models for bearing fault diagnosis using the CWRU dataset: A multi-label approach
Jul 19, 2024Abstract:This paper proposes a novel approach for modeling the problem of fault diagnosis using the Case Western Reserve University (CWRU) bearing fault dataset. Although the dataset is considered a standard reference for testing new algorithms, the typical dataset division suffers from data leakage, as shown by Hendriks et al. (2022) and Abburi et al. (2023), leading to papers reporting over-optimistic results. While their proposed division significantly mitigates this issue, it does not eliminate it entirely. Moreover, their proposed multi-class classification task can still lead to an unrealistic scenario by excluding the possibility of more than one fault type occurring at the same or different locations. As advocated in this paper, a multi-label formulation (detecting the presence of each type of fault for each location) can solve both issues, leading to a scenario closer to reality. Additionally, this approach mitigates the heavy class imbalance of the CWRU dataset, where faulty cases appear much more frequently than healthy cases, even though the opposite is more likely to occur in practice. A multi-label formulation also enables a more precise evaluation using prevalence-independent evaluation metrics for binary classification, such as the ROC curve. Finally, this paper proposes a more realistic dataset division that allows for more diversity in the training dataset while keeping the division free from data leakage. The results show that this new division can significantly improve performance while enabling a fine-grained error analysis. As an application of our approach, a comparative benchmark is performed using several state-of-the-art deep learning models applied to 1D and 2D signal representations in time and/or frequency domains.
Optical Image-to-Image Translation Using Denoising Diffusion Models: Heterogeneous Change Detection as a Use Case
Apr 17, 2024



Abstract:We introduce an innovative deep learning-based method that uses a denoising diffusion-based model to translate low-resolution images to high-resolution ones from different optical sensors while preserving the contents and avoiding undesired artifacts. The proposed method is trained and tested on a large and diverse data set of paired Sentinel-II and Planet Dove images. We show that it can solve serious image generation issues observed when the popular classifier-free guided Denoising Diffusion Implicit Model (DDIM) framework is used in the task of Image-to-Image Translation of multi-sensor optical remote sensing images and that it can generate large images with highly consistent patches, both in colors and in features. Moreover, we demonstrate how our method improves heterogeneous change detection results in two urban areas: Beirut, Lebanon, and Austin, USA. Our contributions are: i) a new training and testing algorithm based on denoising diffusion models for optical image translation; ii) a comprehensive image quality evaluation and ablation study; iii) a comparison with the classifier-free guided DDIM framework; and iv) change detection experiments on heterogeneous data.
Selective Prediction for Semantic Segmentation using Post-Hoc Confidence Estimation and Its Performance under Distribution Shift
Feb 16, 2024Abstract:Semantic segmentation plays a crucial role in various computer vision applications, yet its efficacy is often hindered by the lack of high-quality labeled data. To address this challenge, a common strategy is to leverage models trained on data from different populations, such as publicly available datasets. This approach, however, leads to the distribution shift problem, presenting a reduced performance on the population of interest. In scenarios where model errors can have significant consequences, selective prediction methods offer a means to mitigate risks and reduce reliance on expert supervision. This paper investigates selective prediction for semantic segmentation in low-resource settings, thus focusing on post-hoc confidence estimators applied to pre-trained models operating under distribution shift. We propose a novel image-level confidence measure tailored for semantic segmentation and demonstrate its effectiveness through experiments on three medical imaging tasks. Our findings show that post-hoc confidence estimators offer a cost-effective approach to reducing the impacts of distribution shift.
Does pre-training on brain-related tasks results in better deep-learning-based brain age biomarkers?
Jul 11, 2023Abstract:Brain age prediction using neuroimaging data has shown great potential as an indicator of overall brain health and successful aging, as well as a disease biomarker. Deep learning models have been established as reliable and efficient brain age estimators, being trained to predict the chronological age of healthy subjects. In this paper, we investigate the impact of a pre-training step on deep learning models for brain age prediction. More precisely, instead of the common approach of pre-training on natural imaging classification, we propose pre-training the models on brain-related tasks, which led to state-of-the-art results in our experiments on ADNI data. Furthermore, we validate the resulting brain age biomarker on images of patients with mild cognitive impairment and Alzheimer's disease. Interestingly, our results indicate that better-performing deep learning models in terms of brain age prediction on healthy patients do not result in more reliable biomarkers.
Improving selective classification performance of deep neural networks through post-hoc logit normalization and temperature scaling
May 24, 2023



Abstract:This paper addresses the problem of selective classification for deep neural networks, where a model is allowed to abstain from low-confidence predictions to avoid potential errors. Specifically, we tackle the problem of optimizing the confidence estimator of a fixed classifier, aiming to enhance its misclassification detection performance, i.e., its ability to discriminate between correct and incorrect predictions by assigning higher confidence values to the correct ones. Previous work has found that different classifiers exhibit varying levels of misclassification detection performance, particularly when using the maximum softmax probability (MSP) as a measure of confidence. However, we argue that these findings are mainly due to a sub-optimal confidence estimator being used for each model. To overcome this issue, we propose a simple and efficient post-hoc confidence estimator, named $p$-NormSoftmax, which consists of transforming the logits through $p$-norm normalization and temperature scaling, followed by taking the MSP, where $p$ and the temperature are optimized based on a hold-out set. This estimator can be easily applied on top of an already trained model and, in many cases, can significantly improve its selective classification performance. When applied to 84 pretrained Imagenet classifiers, our method yields an average improvement of 16% in the area under the risk-coverage curve (AURC), exceeding 40% for some models. Furthermore, after applying $p$-NormSoftmax, we observe that these models exhibit approximately the same level of misclassification detection performance, implying that a model's selective classification performance is almost entirely determined by its accuracy at full coverage.
Towards fully automated deep-learning-based brain tumor segmentation: is brain extraction still necessary?
Dec 14, 2022
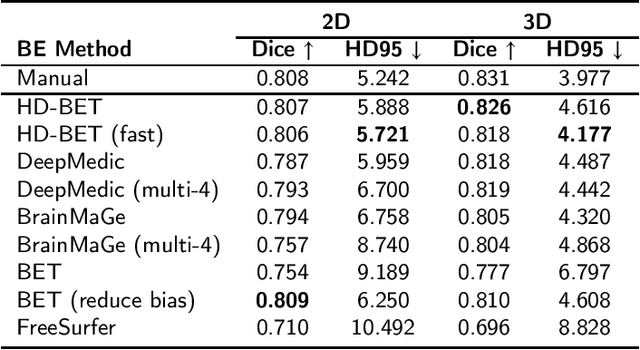

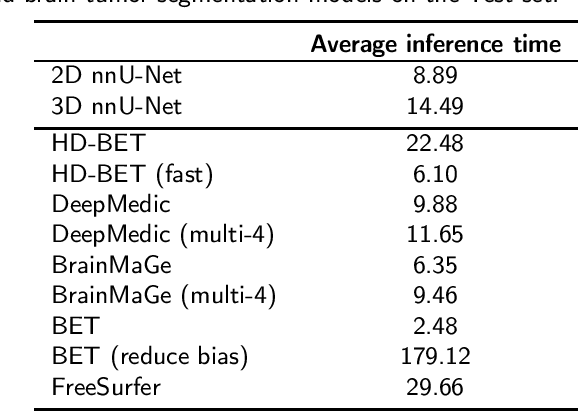
Abstract:State-of-the-art brain tumor segmentation is based on deep learning models applied to multi-modal MRIs. Currently, these models are trained on images after a preprocessing stage that involves registration, interpolation, brain extraction (BE, also known as skull-stripping) and manual correction by an expert. However, for clinical practice, this last step is tedious and time-consuming and, therefore, not always feasible, resulting in skull-stripping faults that can negatively impact the tumor segmentation quality. Still, the extent of this impact has never been measured for any of the many different BE methods available. In this work, we propose an automatic brain tumor segmentation pipeline and evaluate its performance with multiple BE methods. Our experiments show that the choice of a BE method can compromise up to 15.7% of the tumor segmentation performance. Moreover, we propose training and testing tumor segmentation models on non-skull-stripped images, effectively discarding the BE step from the pipeline. Our results show that this approach leads to a competitive performance at a fraction of the time. We conclude that, in contrast to the current paradigm, training tumor segmentation models on non-skull-stripped images can be the best option when high performance in clinical practice is desired.
Predição de Incidência de Lesão por Pressão em Pacientes de UTI usando Aprendizado de Máquina
Dec 23, 2021Abstract:Pressure ulcers have high prevalence in ICU patients but are preventable if identified in initial stages. In practice, the Braden scale is used to classify high-risk patients. This paper investigates the use of machine learning in electronic health records data for this task, by using data available in MIMIC-III v1.4. Two main contributions are made: a new approach for evaluating models that considers all predictions made during a stay, and a new training method for the machine learning models. The results show a superior performance in comparison to the state of the art; moreover, all models surpass the Braden scale in every operating point in the precision-recall curve. -- -- Les\~oes por press\~ao possuem alta preval\^encia em pacientes de UTI e s\~ao preven\'iveis ao serem identificadas em est\'agios iniciais. Na pr\'atica utiliza-se a escala de Braden para classifica\c{c}\~ao de pacientes em risco. Este artigo investiga o uso de aprendizado de m\'aquina em dados de registros eletr\^onicos para este fim, a partir da base de dados MIMIC-III v1.4. S\~ao feitas duas contribui\c{c}\~oes principais: uma nova abordagem para a avalia\c{c}\~ao dos modelos e da escala de Braden levando em conta todas as predi\c{c}\~oes feitas ao longo das interna\c{c}\~oes, e um novo m\'etodo de treinamento para os modelos de aprendizado de m\'aquina. Os resultados obtidos superam o estado da arte e verifica-se que os modelos superam significativamente a escala de Braden em todos os pontos de opera\c{c}\~ao da curva de precis\~ao por sensibilidade.
Predição da Idade Cerebral a partir de Imagens de Ressonância Magnética utilizando Redes Neurais Convolucionais
Dec 23, 2021Abstract:In this work, deep learning techniques for brain age prediction from magnetic resonance images are investigated, aiming to assist in the identification of biomarkers of the natural aging process. The identification of biomarkers is useful for detecting an early-stage neurodegenerative process, as well as for predicting age-related or non-age-related cognitive decline. Two techniques are implemented and compared in this work: a 3D Convolutional Neural Network applied to the volumetric image and a 2D Convolutional Neural Network applied to slices from the axial plane, with subsequent fusion of individual predictions. The best result was obtained by the 2D model, which achieved a mean absolute error of 3.83 years. -- Neste trabalho s\~ao investigadas t\'ecnicas de aprendizado profundo para a predi\c{c}\~ao da idade cerebral a partir de imagens de resson\^ancia magn\'etica, visando auxiliar na identifica\c{c}\~ao de biomarcadores do processo natural de envelhecimento. A identifica\c{c}\~ao de biomarcadores \'e \'util para a detec\c{c}\~ao de um processo neurodegenerativo em est\'agio inicial, al\'em de possibilitar prever um decl\'inio cognitivo relacionado ou n\~ao \`a idade. Duas t\'ecnicas s\~ao implementadas e comparadas neste trabalho: uma Rede Neural Convolucional 3D aplicada na imagem volum\'etrica e uma Rede Neural Convolucional 2D aplicada a fatias do plano axial, com posterior fus\~ao das predi\c{c}\~oes individuais. O melhor resultado foi obtido pelo modelo 2D, que alcan\c{c}ou um erro m\'edio absoluto de 3.83 anos.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge