Daniele Bertolini
Digital Twin Generators for Disease Modeling
May 02, 2024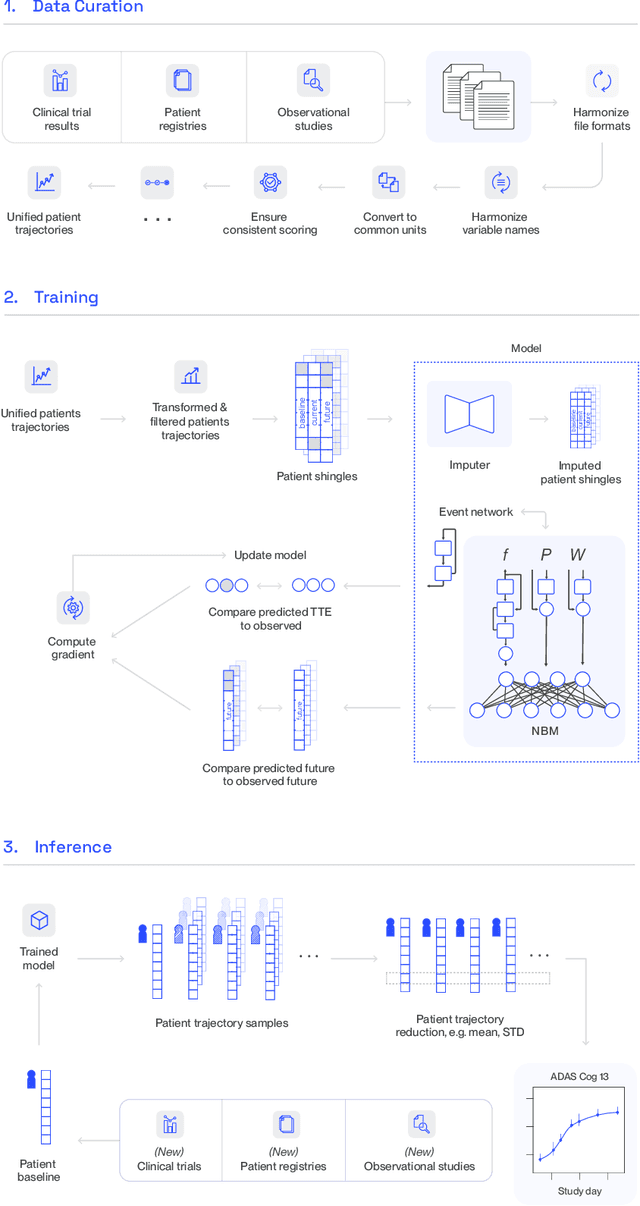
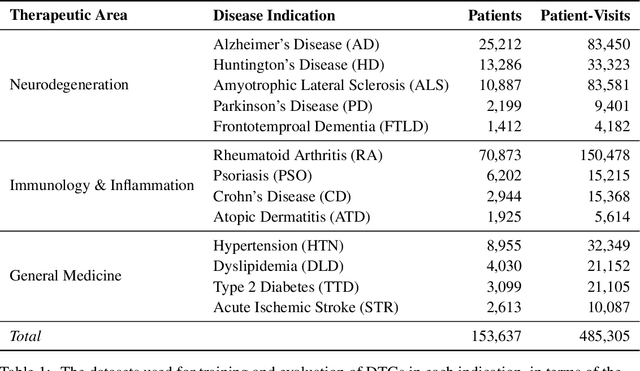
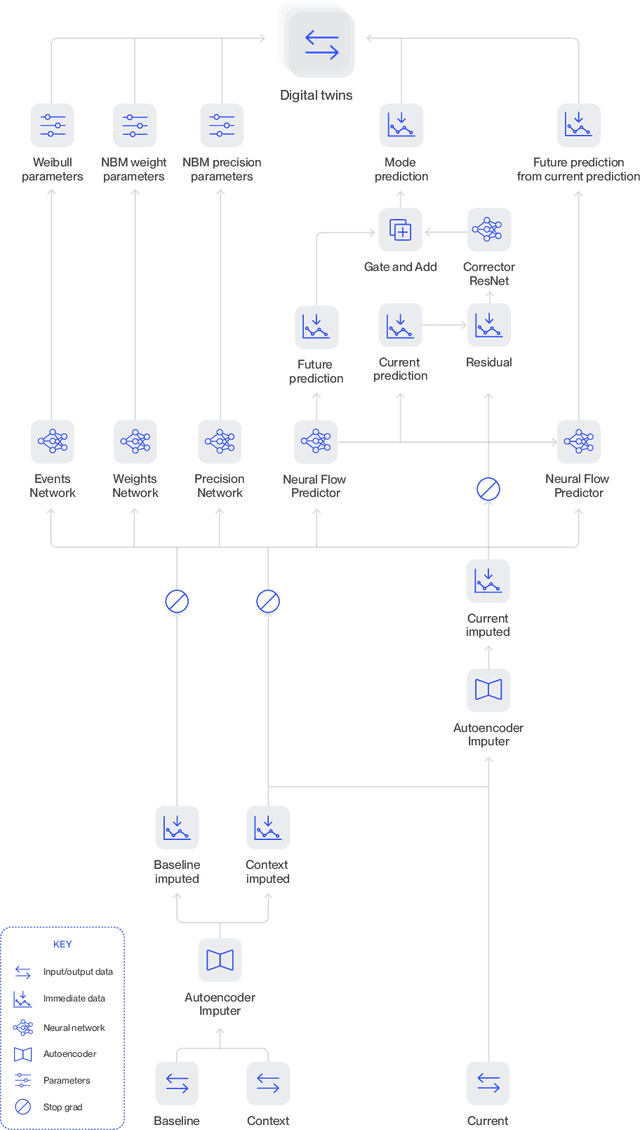
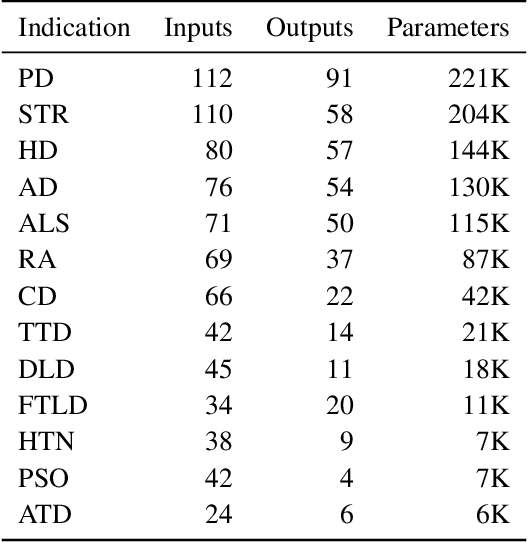
Abstract:A patient's digital twin is a computational model that describes the evolution of their health over time. Digital twins have the potential to revolutionize medicine by enabling individual-level computer simulations of human health, which can be used to conduct more efficient clinical trials or to recommend personalized treatment options. Due to the overwhelming complexity of human biology, machine learning approaches that leverage large datasets of historical patients' longitudinal health records to generate patients' digital twins are more tractable than potential mechanistic models. In this manuscript, we describe a neural network architecture that can learn conditional generative models of clinical trajectories, which we call Digital Twin Generators (DTGs), that can create digital twins of individual patients. We show that the same neural network architecture can be trained to generate accurate digital twins for patients across 13 different indications simply by changing the training set and tuning hyperparameters. By introducing a general purpose architecture, we aim to unlock the ability to scale machine learning approaches to larger datasets and across more indications so that a digital twin could be created for any patient in the world.
Modeling Disease Progression in Mild Cognitive Impairment and Alzheimer's Disease with Digital Twins
Dec 24, 2020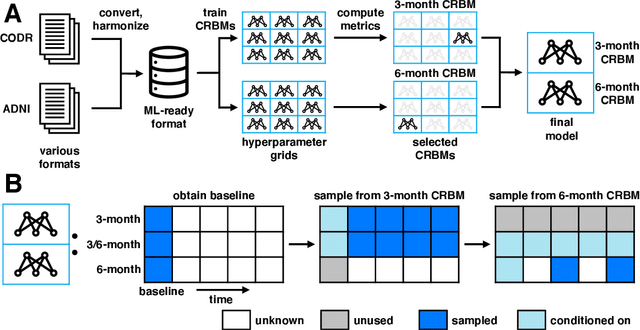

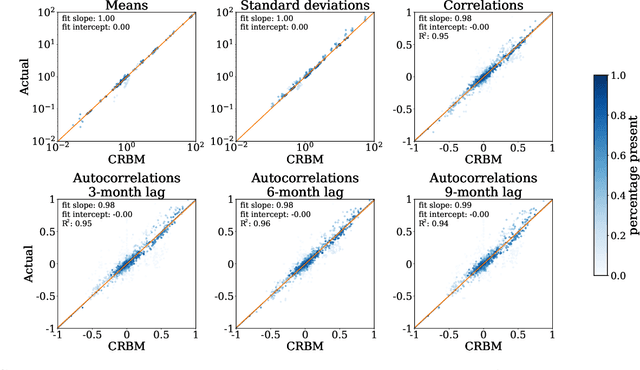
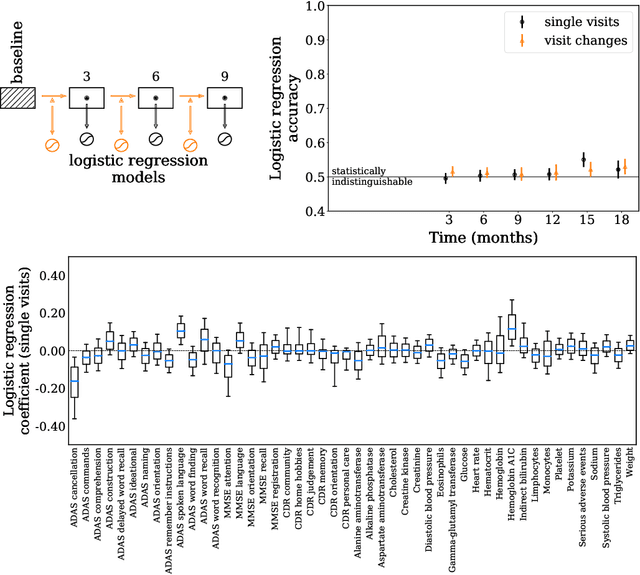
Abstract:Alzheimer's Disease (AD) is a neurodegenerative disease that affects subjects in a broad range of severity and is assessed in clinical trials with multiple cognitive and functional instruments. As clinical trials in AD increasingly focus on earlier stages of the disease, especially Mild Cognitive Impairment (MCI), the ability to model subject outcomes across the disease spectrum is extremely important. We use unsupervised machine learning models called Conditional Restricted Boltzmann Machines (CRBMs) to create Digital Twins of AD subjects. Digital Twins are simulated clinical records that share baseline data with actual subjects and comprehensively model their outcomes under standard-of-care. The CRBMs are trained on a large set of records from subjects in observational studies and the placebo arms of clinical trials across the AD spectrum. These data exhibit a challenging, but common, patchwork of measured and missing observations across subjects in the dataset, and we present a novel model architecture designed to learn effectively from it. We evaluate performance against a held-out test dataset and show how Digital Twins simultaneously capture the progression of a number of key endpoints in clinical trials across a broad spectrum of disease severity, including MCI and mild-to-moderate AD.
Incidence Networks for Geometric Deep Learning
May 27, 2019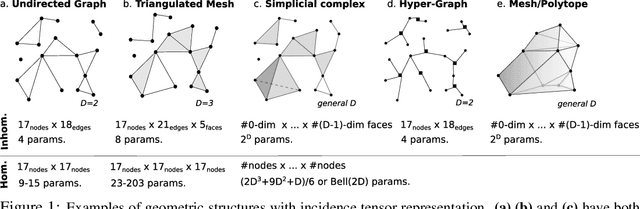



Abstract:One may represent a graph using both its node-edge and its node-node incidence matrices. This choice of representation leads to two classes of equivariant neural networks for attributed graphs that we call incidence networks. Moving beyond graphs, incidence tensors can represent higher dimensional geometric data structures, such as attributed mesh and polytope. For example, a triangulated mesh can be represented using either a "homogeneous" node-node-node or an "inhomogeneous" node-edge-face incidence tensor. This is analogous to the choice of node-node vs. node-edge in graphs. We address the question of "which of these combinations of representation and deep model is more expressive?" and prove that for graphs, homogeneous and inhomogeneous models have equal expressive power. For higher dimensional incidence tensors, we prove that the inhomogeneous model, which is simpler and more practical, can also be more expressive. We demonstrate the effectiveness of incidence networks in quantum chemistry domain by reporting state-of-the-art on QM9 dataset, using both homogeneous and inhomogeneous representations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge