Aaron M. Smith
Modeling Disease Progression in Mild Cognitive Impairment and Alzheimer's Disease with Digital Twins
Dec 24, 2020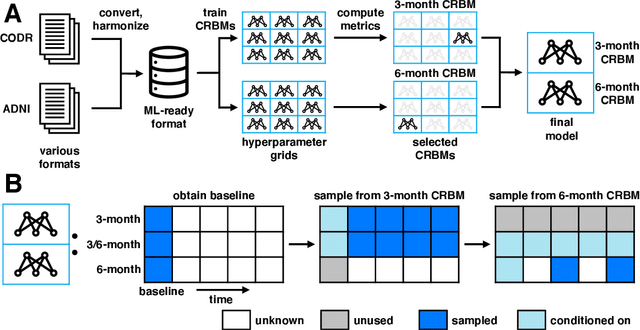

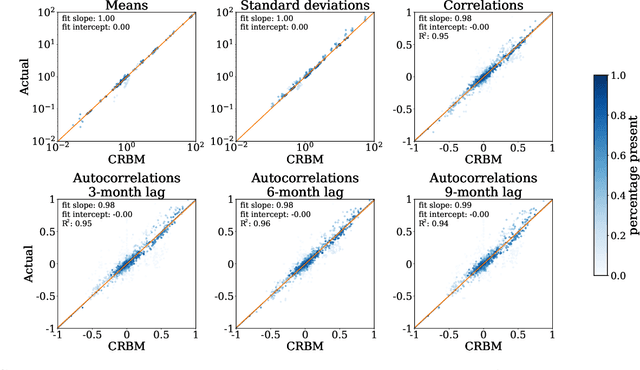
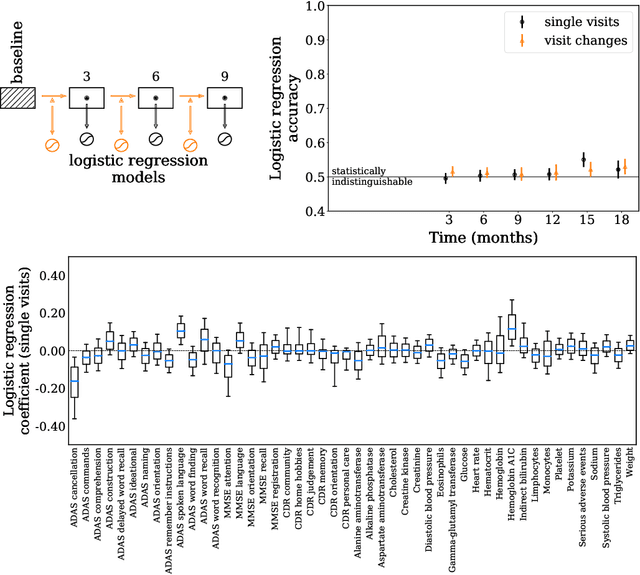
Abstract:Alzheimer's Disease (AD) is a neurodegenerative disease that affects subjects in a broad range of severity and is assessed in clinical trials with multiple cognitive and functional instruments. As clinical trials in AD increasingly focus on earlier stages of the disease, especially Mild Cognitive Impairment (MCI), the ability to model subject outcomes across the disease spectrum is extremely important. We use unsupervised machine learning models called Conditional Restricted Boltzmann Machines (CRBMs) to create Digital Twins of AD subjects. Digital Twins are simulated clinical records that share baseline data with actual subjects and comprehensively model their outcomes under standard-of-care. The CRBMs are trained on a large set of records from subjects in observational studies and the placebo arms of clinical trials across the AD spectrum. These data exhibit a challenging, but common, patchwork of measured and missing observations across subjects in the dataset, and we present a novel model architecture designed to learn effectively from it. We evaluate performance against a held-out test dataset and show how Digital Twins simultaneously capture the progression of a number of key endpoints in clinical trials across a broad spectrum of disease severity, including MCI and mild-to-moderate AD.
Generating Digital Twins with Multiple Sclerosis Using Probabilistic Neural Networks
Feb 04, 2020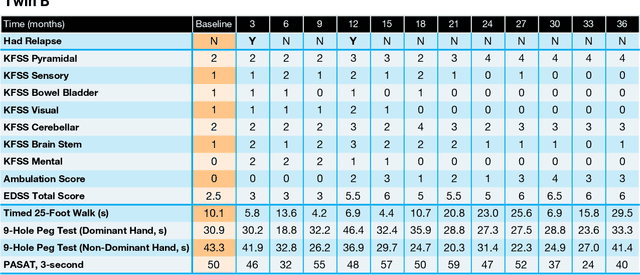
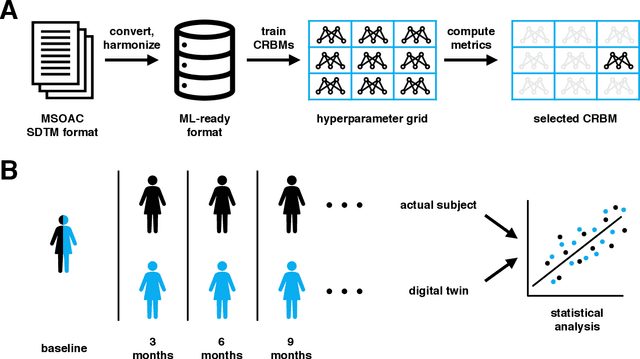
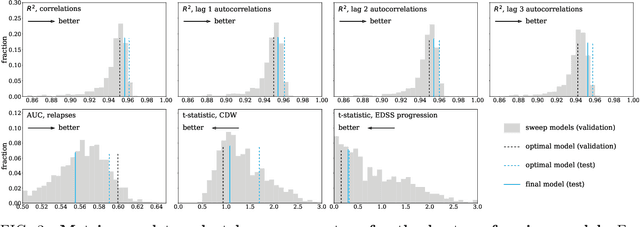
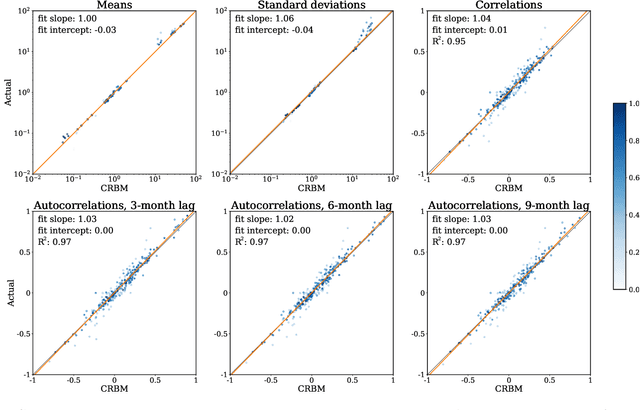
Abstract:Multiple Sclerosis (MS) is a neurodegenerative disorder characterized by a complex set of clinical assessments. We use an unsupervised machine learning model called a Conditional Restricted Boltzmann Machine (CRBM) to learn the relationships between covariates commonly used to characterize subjects and their disease progression in MS clinical trials. A CRBM is capable of generating digital twins, which are simulated subjects having the same baseline data as actual subjects. Digital twins allow for subject-level statistical analyses of disease progression. The CRBM is trained using data from 2395 subjects enrolled in the placebo arms of clinical trials across the three primary subtypes of MS. We discuss how CRBMs are trained and show that digital twins generated by the model are statistically indistinguishable from their actual subject counterparts along a number of measures.
Using deep learning for comprehensive, personalized forecasting of Alzheimer's Disease progression
Jul 10, 2018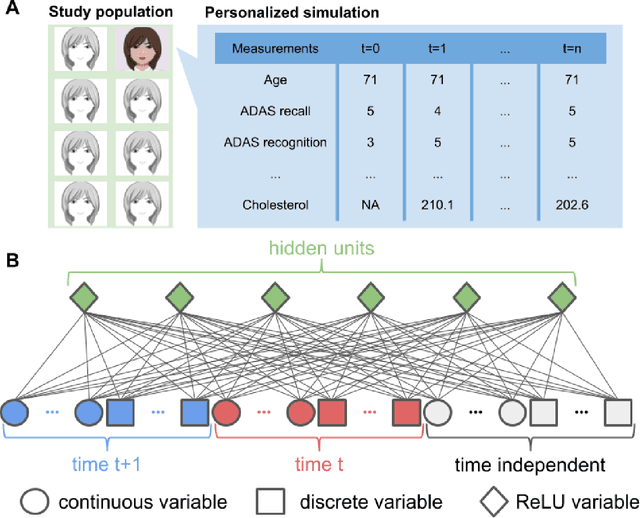
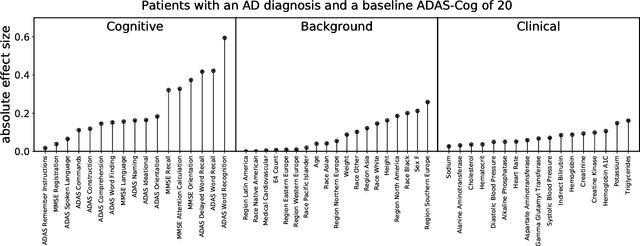
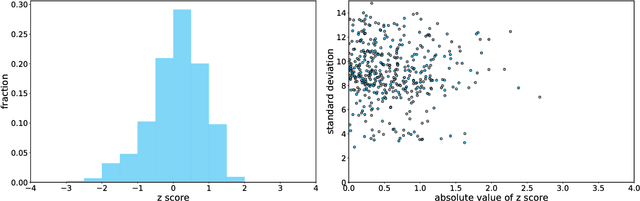
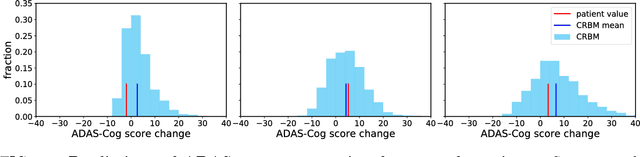
Abstract:A patient is more than one number, yet most approaches to machine learning from electronic health data can only predict a single endpoint. Here, we present an alternative -- using unsupervised deep learning to simulate detailed patient trajectories. We use data comprising 18-month longitudinal trajectories of 42 clinical variables from 1908 patients with Mild Cognitive Impairment (MCI) or Alzheimer's Disease (AD) to train a model for personalized forecasting of disease progression. Our model simulates the evolution of each sub-component of cognitive exams, laboratory tests, and their associations with baseline clinical characteristics, generating both predictions and their confidence intervals. Even though it is not trained to predict changes in disease severity, our unsupervised model predicts changes in total ADAS-Cog scores with the same accuracy as specifically trained supervised models. We show how simulations can be used to interpret our model and demonstrate how to create synthetic control arm data for AD clinical trials. Our model's ability to simultaneously predict dozens of characteristics of a patient at any point in the future is a crucial step forward in computational precision medicine.
Boltzmann Encoded Adversarial Machines
Apr 23, 2018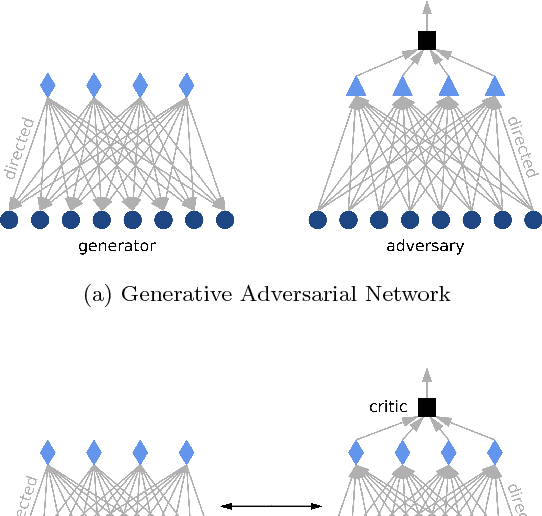
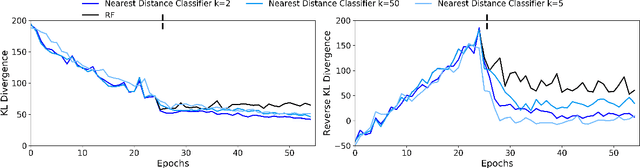
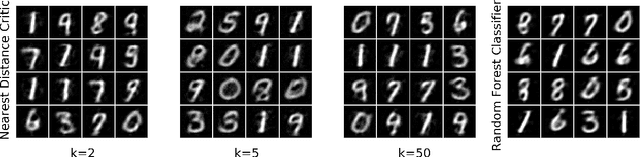
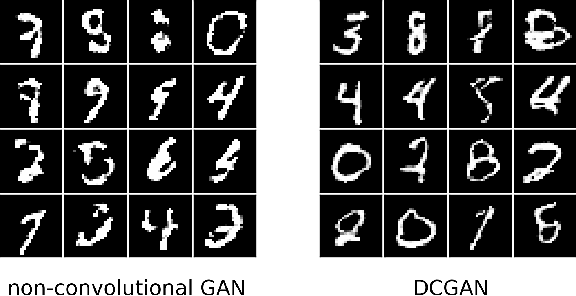
Abstract:Restricted Boltzmann Machines (RBMs) are a class of generative neural network that are typically trained to maximize a log-likelihood objective function. We argue that likelihood-based training strategies may fail because the objective does not sufficiently penalize models that place a high probability in regions where the training data distribution has low probability. To overcome this problem, we introduce Boltzmann Encoded Adversarial Machines (BEAMs). A BEAM is an RBM trained against an adversary that uses the hidden layer activations of the RBM to discriminate between the training data and the probability distribution generated by the model. We present experiments demonstrating that BEAMs outperform RBMs and GANs on multiple benchmarks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge