Daniel S. Zisook
Task formulation for Extracting Social Determinants of Health from Clinical Narratives
Jan 26, 2023
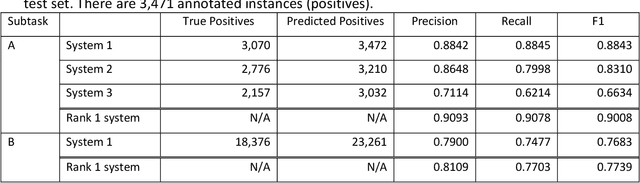
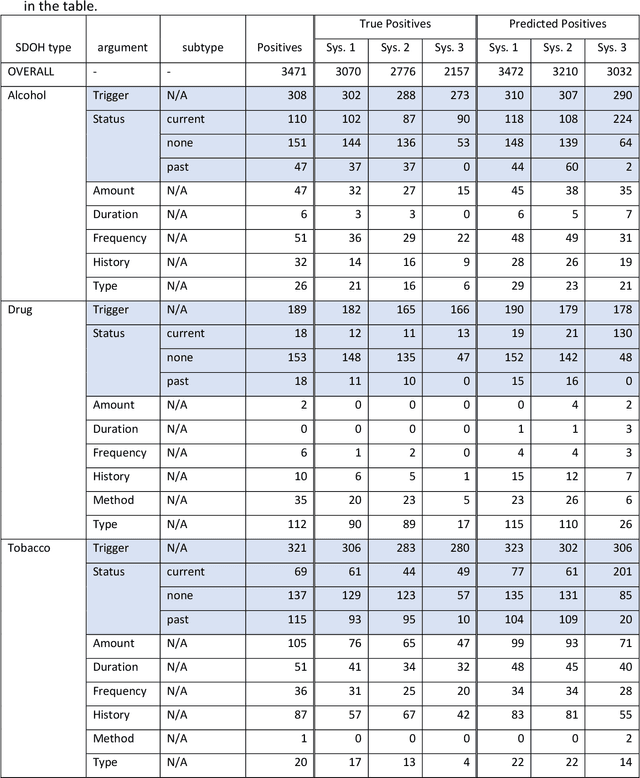
Abstract:Objective: The 2022 n2c2 NLP Challenge posed identification of social determinants of health (SDOH) in clinical narratives. We present three systems that we developed for the Challenge and discuss the distinctive task formulation used in each of the three systems. Materials and Methods: The first system identifies target pieces of information independently using machine learning classifiers. The second system uses a large language model (LLM) to extract complete structured outputs per document. The third system extracts candidate phrases using machine learning and identifies target relations with hand-crafted rules. Results: The three systems achieved F1 scores of 0.884, 0.831, and 0.663 in the Subtask A of the Challenge, which are ranked third, seventh, and eighth among the 15 participating teams. The review of the extraction results from our systems reveals characteristics of each approach and those of the SODH extraction task. Discussion: Phrases and relations annotated in the task is unique and diverse, not conforming to the conventional event extraction task. These annotations are difficult to model with limited training data. The system that extracts information independently, ignoring the annotated relations, achieves the highest F1 score. Meanwhile, LLM with its versatile capability achieves the high F1 score, while respecting the annotated relations. The rule-based system tackling relation extraction obtains the low F1 score, while it is the most explainable approach. Conclusion: The F1 scores of the three systems vary in this challenge setting, but each approach has advantages and disadvantages in a practical application. The selection of the approach depends not only on the F1 score but also on the requirements in the application.
SmartTriage: A system for personalized patient data capture, documentation generation, and decision support
Oct 19, 2020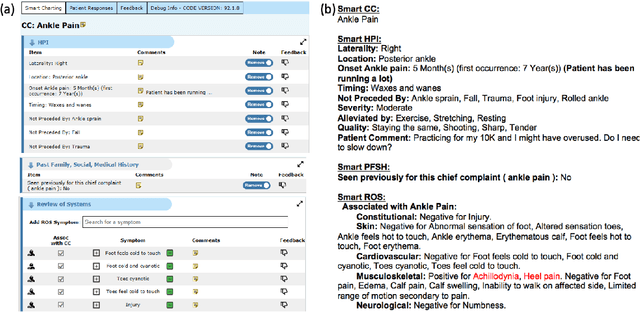
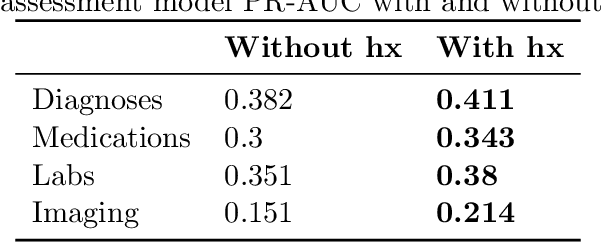
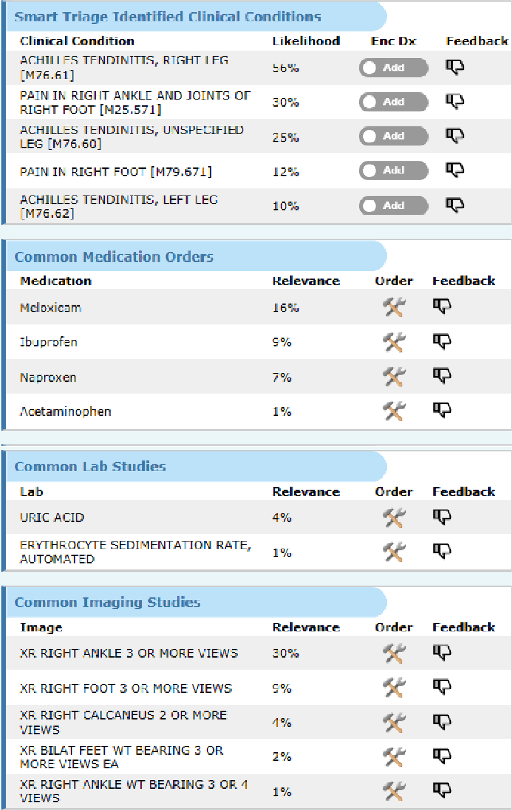
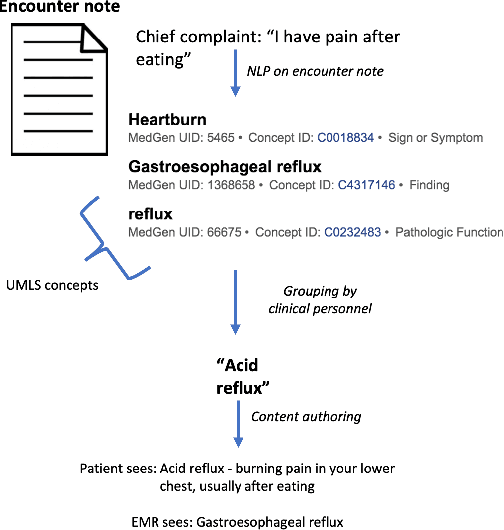
Abstract:Symptom checkers have emerged as an important tool for collecting symptoms and diagnosing patients, minimizing the involvement of clinical personnel. We developed a machine-learning-backed system, SmartTriage, which goes beyond conventional symptom checking through a tight bi-directional integration with the electronic medical record (EMR). Conditioned on EMR-derived patient history, our system identifies the patient's chief complaint from a free-text entry and then asks a series of discrete questions to obtain relevant symptomatology. The patient-specific data are used to predict detailed ICD-10-CM codes as well as medication, laboratory, and imaging orders. Patient responses and clinical decision support (CDS) predictions are then inserted back into the EMR. To train the machine learning components of SmartTriage, we employed novel data sets of over 25 million primary care encounters and 1 million patient free-text reason-for-visit entries. These data sets were used to construct: (1) a long short-term memory (LSTM) based patient history representation, (2) a fine-tuned transformer model for chief complaint extraction, (3) a random forest model for question sequencing, and (4) a feed-forward network for CDS predictions. We also present the full production architecture for the pilot deployment of SmartTriage that covers 337 patient chief complaints.
Evaluating robustness of language models for chief complaint extraction from patient-generated text
Nov 15, 2019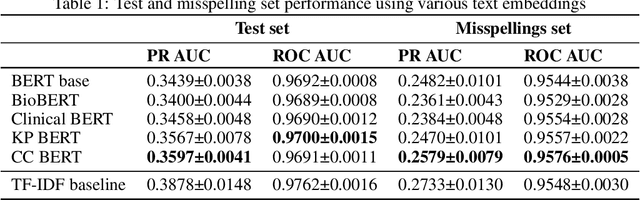
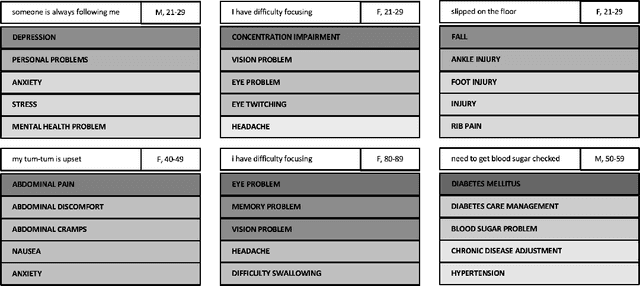
Abstract:Automated classification of chief complaints from patient-generated text is a critical first step in developing scalable platforms to triage patients without human intervention. In this work, we evaluate several approaches to chief complaint classification using a novel Chief Complaint (CC) Dataset that contains ~200,000 patient-generated reasons-for-visit entries mapped to a set of 795 discrete chief complaints. We examine the use of several fine-tuned bidirectional transformer (BERT) models trained on both unrelated texts as well as on the CC dataset. We contrast this performance with a TF-IDF baseline. Our evaluation has three components: (1) a random test hold-out from the original dataset; (2) a "misspelling set," consisting of a hand-selected subset of the test set, where every entry has at least one misspelling; (3) a separate experimenter-generated free-text set. We find that the TF-IDF model performs significantly better than the strongest BERT-based model on the test (best BERT PR-AUC $0.3597 \pm 0.0041$ vs TF-IDF PR-AUC $0.3878 \pm 0.0148$, $p=7\cdot 10^{-5}$), and is statistically comparable to the misspelling sets (best BERT PR-AUC $0.2579 \pm 0.0079$ vs TF-IDF PR-AUC $0.2733 \pm 0.0130$, $p=0.06$). However, when examining model predictions on experimenter-generated queries, some concerns arise about TF-IDF baseline's robustness. Our results suggest that in certain tasks, simple language embedding baselines may be very performant; however, truly understanding their robustness requires further analysis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge