Manabu Torii
Task formulation for Extracting Social Determinants of Health from Clinical Narratives
Jan 26, 2023
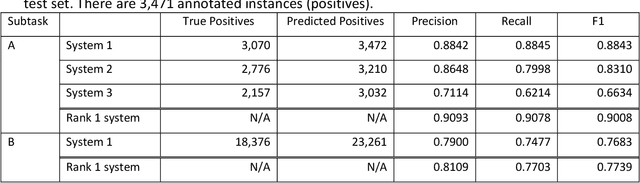
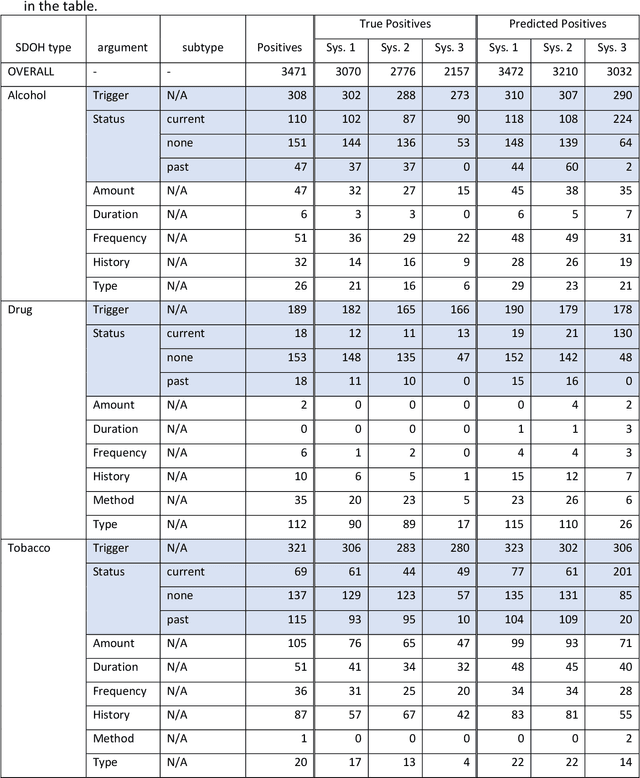
Abstract:Objective: The 2022 n2c2 NLP Challenge posed identification of social determinants of health (SDOH) in clinical narratives. We present three systems that we developed for the Challenge and discuss the distinctive task formulation used in each of the three systems. Materials and Methods: The first system identifies target pieces of information independently using machine learning classifiers. The second system uses a large language model (LLM) to extract complete structured outputs per document. The third system extracts candidate phrases using machine learning and identifies target relations with hand-crafted rules. Results: The three systems achieved F1 scores of 0.884, 0.831, and 0.663 in the Subtask A of the Challenge, which are ranked third, seventh, and eighth among the 15 participating teams. The review of the extraction results from our systems reveals characteristics of each approach and those of the SODH extraction task. Discussion: Phrases and relations annotated in the task is unique and diverse, not conforming to the conventional event extraction task. These annotations are difficult to model with limited training data. The system that extracts information independently, ignoring the annotated relations, achieves the highest F1 score. Meanwhile, LLM with its versatile capability achieves the high F1 score, while respecting the annotated relations. The rule-based system tackling relation extraction obtains the low F1 score, while it is the most explainable approach. Conclusion: The F1 scores of the three systems vary in this challenge setting, but each approach has advantages and disadvantages in a practical application. The selection of the approach depends not only on the F1 score but also on the requirements in the application.
Using natural language processing to extract health-related causality from Twitter messages
Nov 15, 2019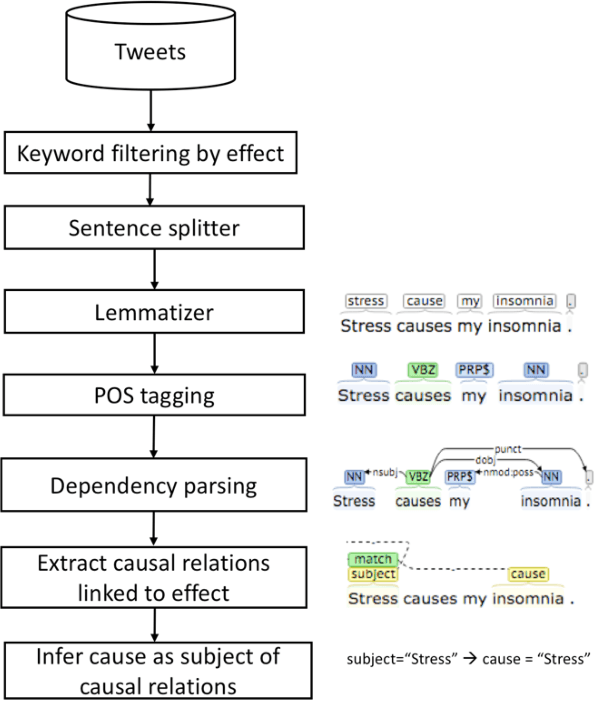
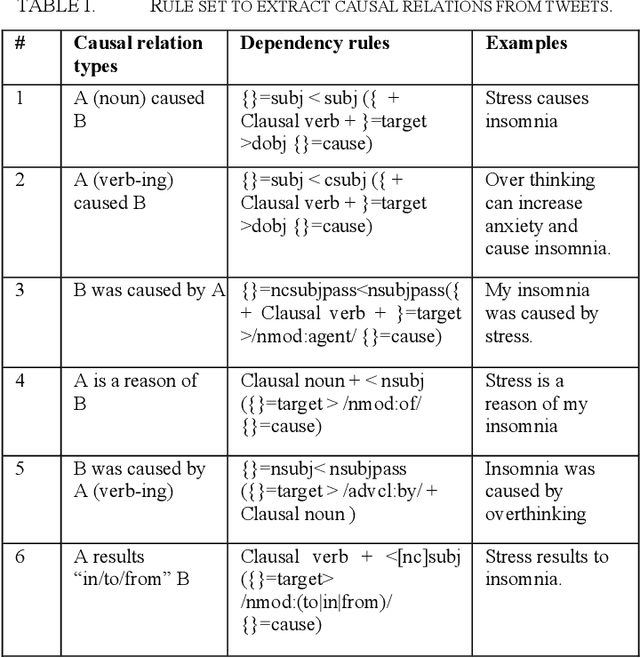
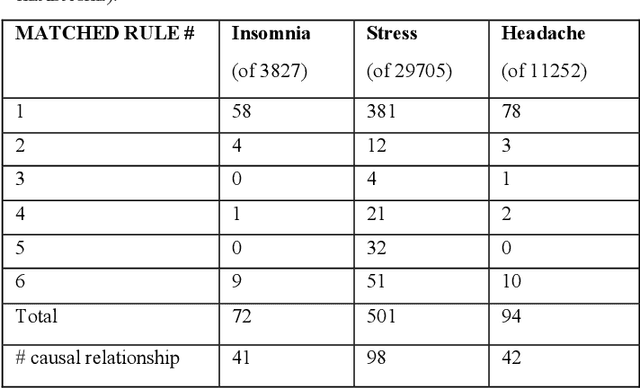
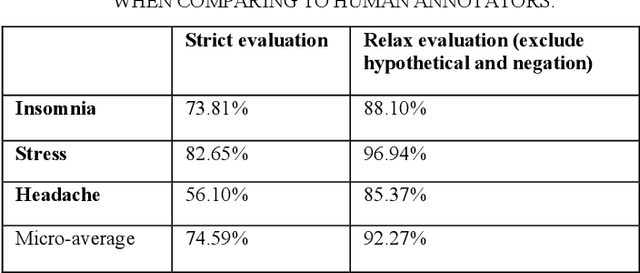
Abstract:Twitter messages (tweets) contain various types of information, which include health-related information. Analysis of health-related tweets would help us understand health conditions and concerns encountered in our daily life. In this work, we evaluated an approach to extracting causal relations from tweets using natural language processing (NLP) techniques. We focused on three health-related topics: stress", "insomnia", and "headache". We proposed a set of lexico-syntactic patterns based on dependency parser outputs to extract causal information. A large dataset consisting of 24 million tweets were used. The results show that our approach achieved an average precision between 74.59% and 92.27%. Analysis of extracted relations revealed interesting findings about health-related in Twitter.
* 5 pages
Rapid Adaptation of POS Tagging for Domain Specific Uses
Oct 31, 2014Abstract:Part-of-speech (POS) tagging is a fundamental component for performing natural language tasks such as parsing, information extraction, and question answering. When POS taggers are trained in one domain and applied in significantly different domains, their performance can degrade dramatically. We present a methodology for rapid adaptation of POS taggers to new domains. Our technique is unsupervised in that a manually annotated corpus for the new domain is not necessary. We use suffix information gathered from large amounts of raw text as well as orthographic information to increase the lexical coverage. We present an experiment in the Biological domain where our POS tagger achieves results comparable to POS taggers specifically trained to this domain.
* 2 pages, 2 tables; appeared in Proceedings of the HLT-NAACL BioNLP Workshop on Linking Natural Language and Biology, June 2006
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge