Daniel M. Faissol
Symbolic Regression via Neural-Guided Genetic Programming Population Seeding
Nov 17, 2021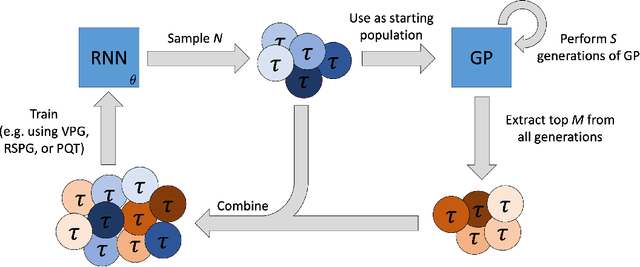
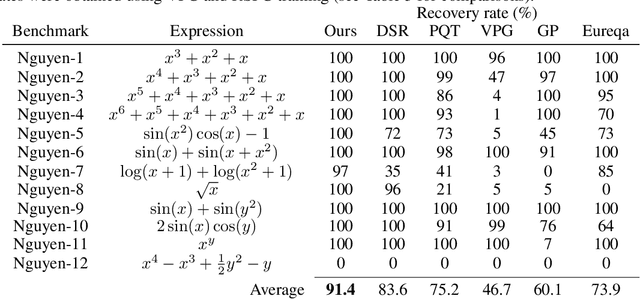
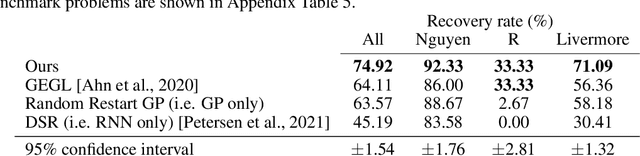
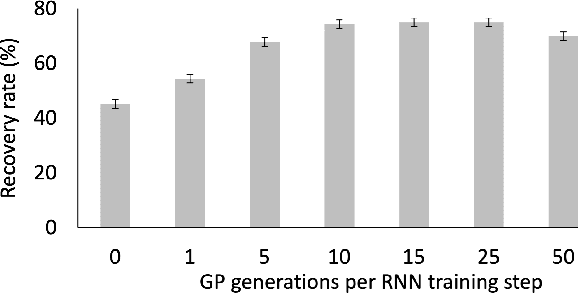
Abstract:Symbolic regression is the process of identifying mathematical expressions that fit observed output from a black-box process. It is a discrete optimization problem generally believed to be NP-hard. Prior approaches to solving the problem include neural-guided search (e.g. using reinforcement learning) and genetic programming. In this work, we introduce a hybrid neural-guided/genetic programming approach to symbolic regression and other combinatorial optimization problems. We propose a neural-guided component used to seed the starting population of a random restart genetic programming component, gradually learning better starting populations. On a number of common benchmark tasks to recover underlying expressions from a dataset, our method recovers 65% more expressions than a recently published top-performing model using the same experimental setup. We demonstrate that running many genetic programming generations without interdependence on the neural-guided component performs better for symbolic regression than alternative formulations where the two are more strongly coupled. Finally, we introduce a new set of 22 symbolic regression benchmark problems with increased difficulty over existing benchmarks. Source code is provided at www.github.com/brendenpetersen/deep-symbolic-optimization.
Improving exploration in policy gradient search: Application to symbolic optimization
Jul 19, 2021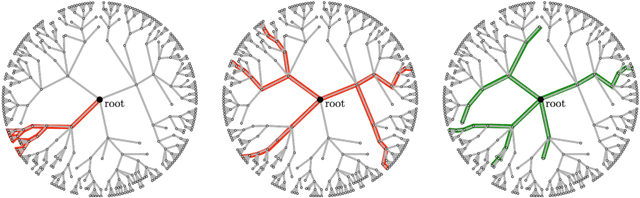
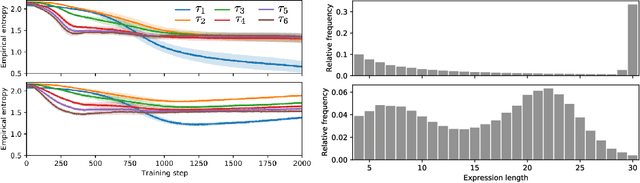
Abstract:Many machine learning strategies designed to automate mathematical tasks leverage neural networks to search large combinatorial spaces of mathematical symbols. In contrast to traditional evolutionary approaches, using a neural network at the core of the search allows learning higher-level symbolic patterns, providing an informed direction to guide the search. When no labeled data is available, such networks can still be trained using reinforcement learning. However, we demonstrate that this approach can suffer from an early commitment phenomenon and from initialization bias, both of which limit exploration. We present two exploration methods to tackle these issues, building upon ideas of entropy regularization and distribution initialization. We show that these techniques can improve the performance, increase sample efficiency, and lower the complexity of solutions for the task of symbolic regression.
* Published in 1st Mathematical Reasoning in General Artificial Intelligence Workshop, ICLR 2021
Precision medicine as a control problem: Using simulation and deep reinforcement learning to discover adaptive, personalized multi-cytokine therapy for sepsis
Feb 08, 2018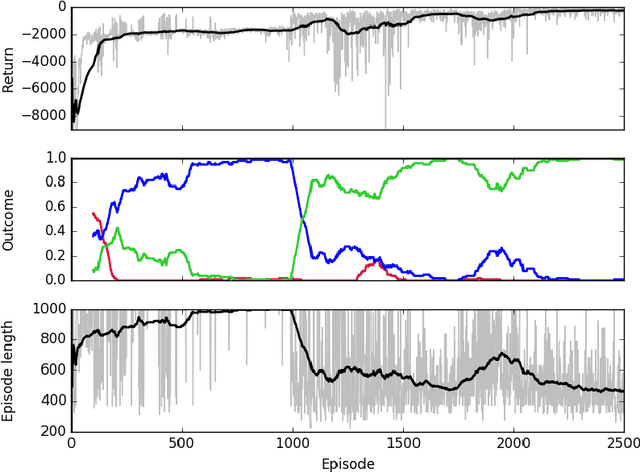
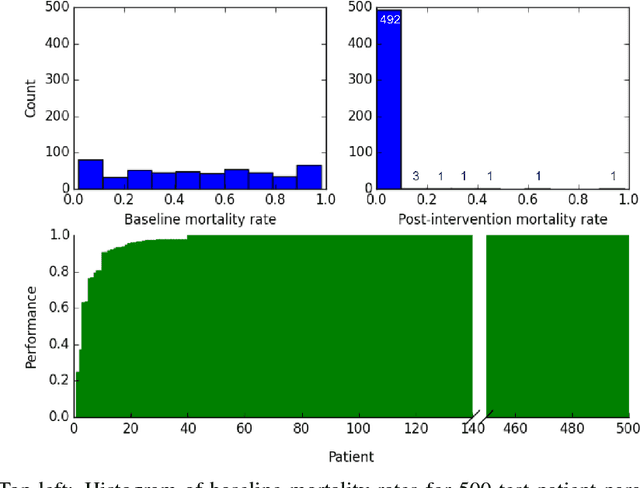
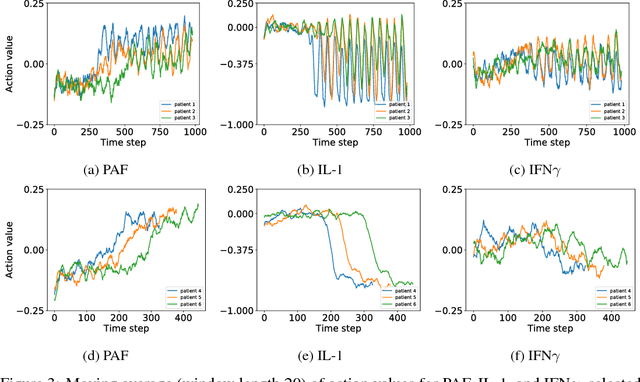
Abstract:Sepsis is a life-threatening condition affecting one million people per year in the US in which dysregulation of the body's own immune system causes damage to its tissues, resulting in a 28 - 50% mortality rate. Clinical trials for sepsis treatment over the last 20 years have failed to produce a single currently FDA approved drug treatment. In this study, we attempt to discover an effective cytokine mediation treatment strategy for sepsis using a previously developed agent-based model that simulates the innate immune response to infection: the Innate Immune Response agent-based model (IIRABM). Previous attempts at reducing mortality with multi-cytokine mediation using the IIRABM have failed to reduce mortality across all patient parameterizations and motivated us to investigate whether adaptive, personalized multi-cytokine mediation can control the trajectory of sepsis and lower patient mortality. We used the IIRABM to compute a treatment policy in which systemic patient measurements are used in a feedback loop to inform future treatment. Using deep reinforcement learning, we identified a policy that achieves 0% mortality on the patient parameterization on which it was trained. More importantly, this policy also achieves 0.8% mortality over 500 randomly selected patient parameterizations with baseline mortalities ranging from 1 - 99% (with an average of 49%) spanning the entire clinically plausible parameter space of the IIRABM. These results suggest that adaptive, personalized multi-cytokine mediation therapy could be a promising approach for treating sepsis. We hope that this work motivates researchers to consider such an approach as part of future clinical trials. To the best of our knowledge, this work is the first to consider adaptive, personalized multi-cytokine mediation therapy for sepsis, and is the first to exploit deep reinforcement learning on a biological simulation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge