Daniel J. Saunders
Minibatch Processing in Spiking Neural Networks
Sep 05, 2019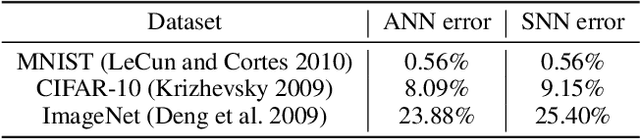
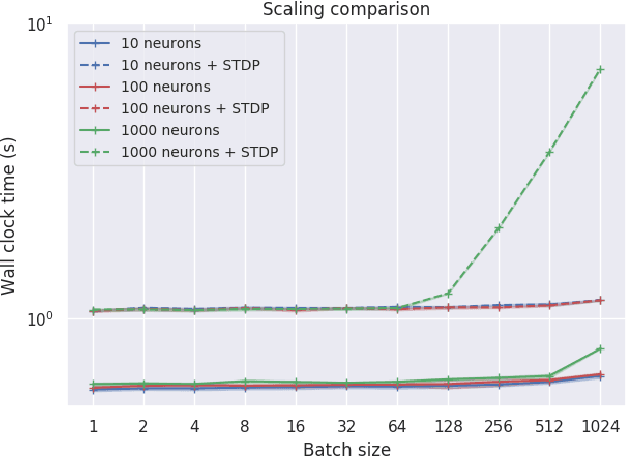

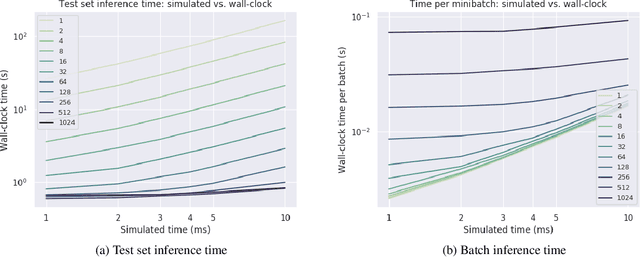
Abstract:Spiking neural networks (SNNs) are a promising candidate for biologically-inspired and energy efficient computation. However, their simulation is notoriously time consuming, and may be seen as a bottleneck in developing competitive training methods with potential deployment on neuromorphic hardware platforms. To address this issue, we provide an implementation of mini-batch processing applied to clock-based SNN simulation, leading to drastically increased data throughput. To our knowledge, this is the first general-purpose implementation of mini-batch processing in a spiking neural networks simulator, which works with arbitrary neuron and synapse models. We demonstrate nearly constant-time scaling with batch size on a simulation setup (up to GPU memory limits), and showcase the effectiveness of large batch sizes in two SNN application domains, resulting in $\approx$880X and $\approx$24X reductions in wall-clock time respectively. Different parameter reduction techniques are shown to produce different learning outcomes in a simulation of networks trained with spike-timing-dependent plasticity. Machine learning practitioners and biological modelers alike may benefit from the drastically reduced simulation time and increased iteration speed this method enables. Code to reproduce the benchmarks and experimental findings in this paper can be found at https://github.com/djsaunde/snn-minibatch.
Lattice Map Spiking Neural Networks (LM-SNNs) for Clustering and Classifying Image Data
Jun 04, 2019
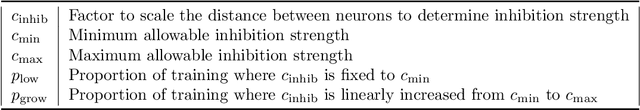
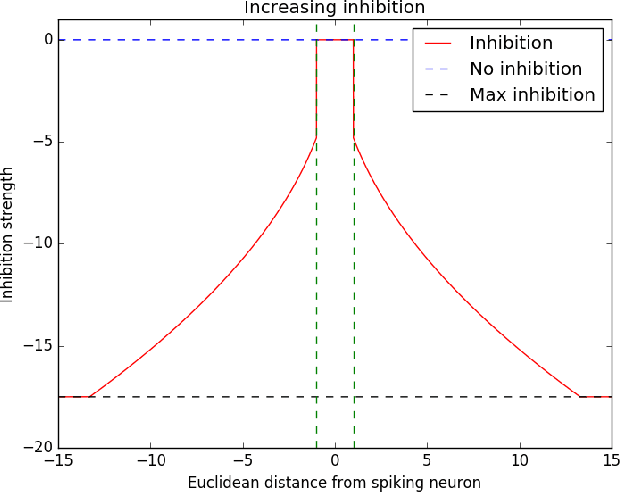
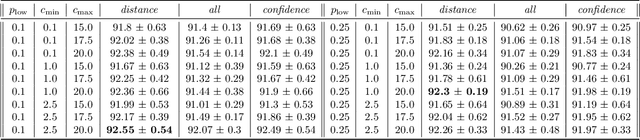
Abstract:Spiking neural networks (SNNs) with a lattice architecture are introduced in this work, combining several desirable properties of SNNs and self-organized maps (SOMs). Networks are trained with biologically motivated, unsupervised learning rules to obtain a self-organized grid of filters via cooperative and competitive excitatory-inhibitory interactions. Several inhibition strategies are developed and tested, such as (i) incrementally increasing inhibition level over the course of network training, and (ii) switching the inhibition level from low to high (two-level) after an initial training segment. During the labeling phase, the spiking activity generated by data with known labels is used to assign neurons to categories of data, which are then used to evaluate the network's classification ability on a held-out set of test data. Several biologically plausible evaluation rules are proposed and compared, including a population-level confidence rating, and an $n$-gram inspired method. The effectiveness of the proposed self-organized learning mechanism is tested using the MNIST benchmark dataset, as well as using images produced by playing the Atari Breakout game.
Locally Connected Spiking Neural Networks for Unsupervised Feature Learning
Apr 12, 2019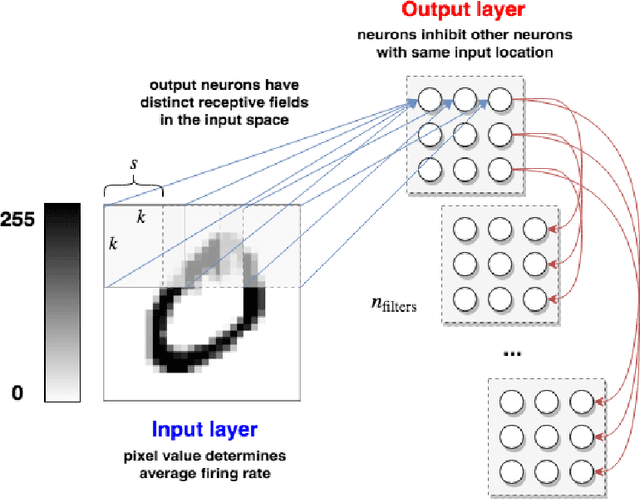
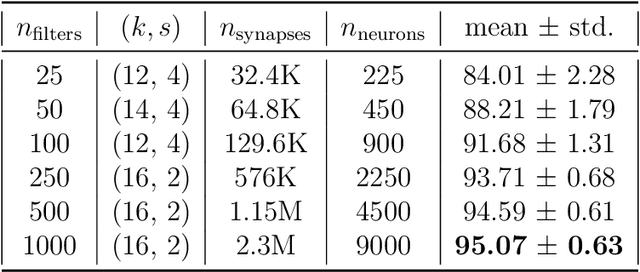
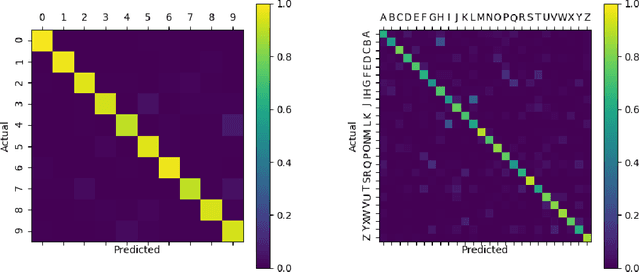
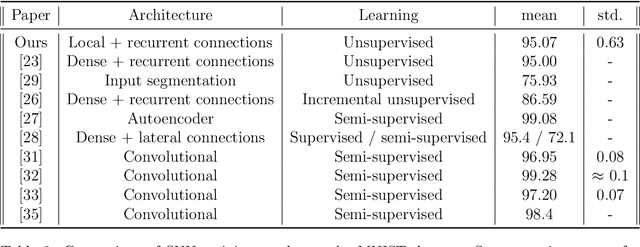
Abstract:In recent years, Spiking Neural Networks (SNNs) have demonstrated great successes in completing various Machine Learning tasks. We introduce a method for learning image features by \textit{locally connected layers} in SNNs using spike-timing-dependent plasticity (STDP) rule. In our approach, sub-networks compete via competitive inhibitory interactions to learn features from different locations of the input space. These \textit{Locally-Connected SNNs} (LC-SNNs) manifest key topological features of the spatial interaction of biological neurons. We explore biologically inspired n-gram classification approach allowing parallel processing over various patches of the the image space. We report the classification accuracy of simple two-layer LC-SNNs on two image datasets, which match the state-of-art performance and are the first results to date. LC-SNNs have the advantage of fast convergence to a dataset representation, and they require fewer learnable parameters than other SNN approaches with unsupervised learning. Robustness tests demonstrate that LC-SNNs exhibit graceful degradation of performance despite the random deletion of large amounts of synapses and neurons.
Improved robustness of reinforcement learning policies upon conversion to spiking neuronal network platforms applied to ATARI games
Mar 26, 2019
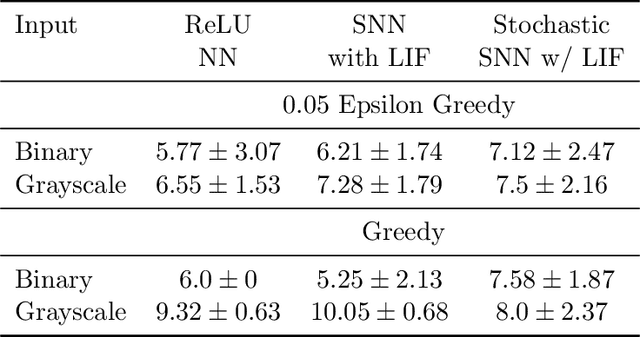
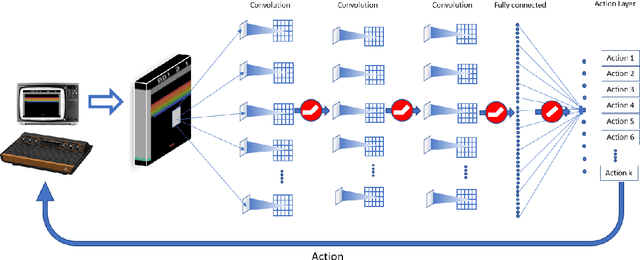
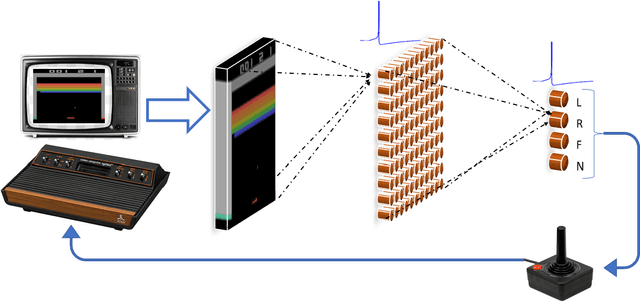
Abstract:Various implementations of Deep Reinforcement Learning (RL) demonstrated excellent performance on tasks that can be solved by trained policy, but they are not without drawbacks. Deep RL suffers from high sensitivity to noisy and missing input and adversarial attacks. To mitigate these deficiencies of deep RL solutions, we suggest involving spiking neural networks (SNNs). Previous work has shown that standard Neural Networks trained using supervised learning for image classification can be converted to SNNs with negligible deterioration in performance. In this paper, we convert Q-Learning ReLU-Networks (ReLU-N) trained using reinforcement learning into SNN. We provide a proof of concept for the conversion of ReLU-N to SNN demonstrating improved robustness to occlusion and better generalization than the original ReLU-N. Moreover, we show promising initial results with converting full-scale Deep Q-networks to SNNs, paving the way for future research.
STDP Learning of Image Patches with Convolutional Spiking Neural Networks
Aug 24, 2018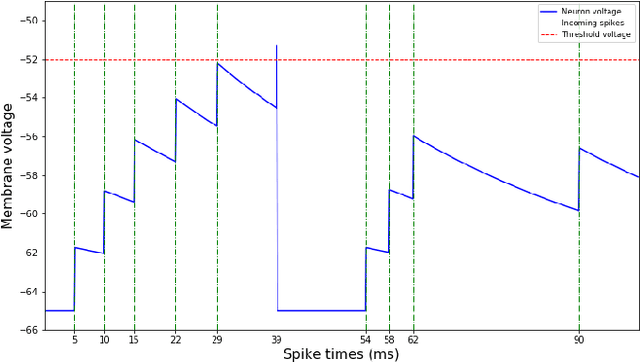
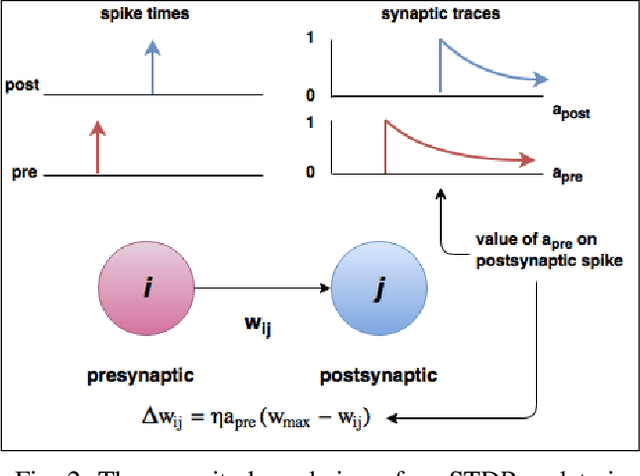
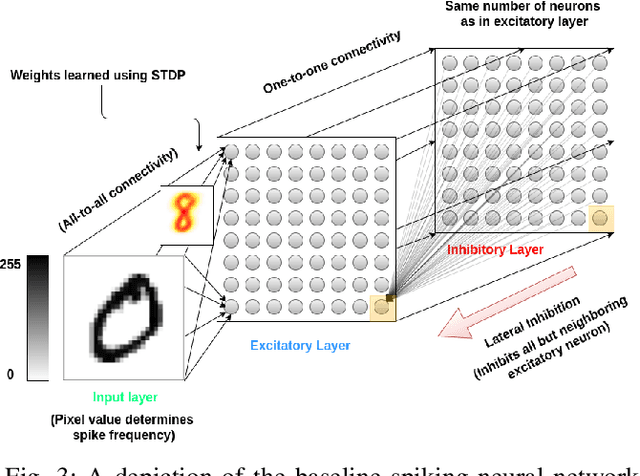
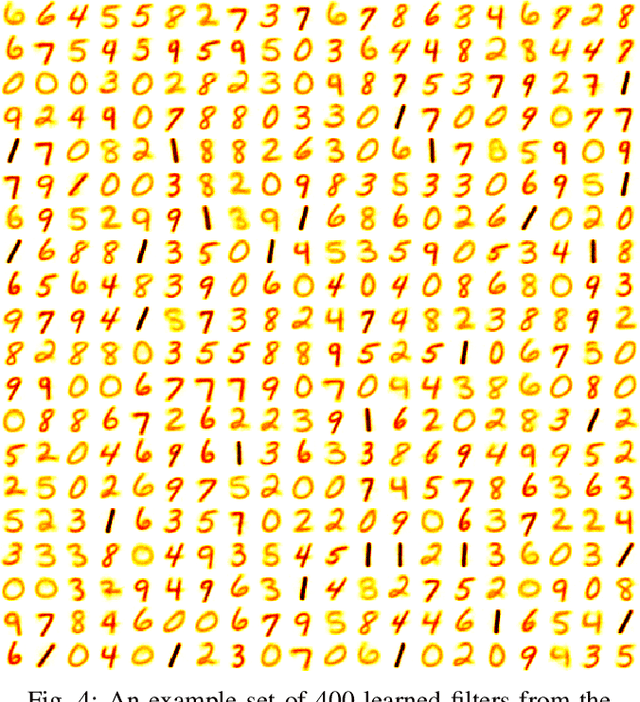
Abstract:Spiking neural networks are motivated from principles of neural systems and may possess unexplored advantages in the context of machine learning. A class of \textit{convolutional spiking neural networks} is introduced, trained to detect image features with an unsupervised, competitive learning mechanism. Image features can be shared within subpopulations of neurons, or each may evolve independently to capture different features in different regions of input space. We analyze the time and memory requirements of learning with and operating such networks. The MNIST dataset is used as an experimental testbed, and comparisons are made between the performance and convergence speed of a baseline spiking neural network.
Unsupervised Learning with Self-Organizing Spiking Neural Networks
Jul 24, 2018



Abstract:We present a system comprising a hybridization of self-organized map (SOM) properties with spiking neural networks (SNNs) that retain many of the features of SOMs. Networks are trained in an unsupervised manner to learn a self-organized lattice of filters via excitatory-inhibitory interactions among populations of neurons. We develop and test various inhibition strategies, such as growing with inter-neuron distance and two distinct levels of inhibition. The quality of the unsupervised learning algorithm is evaluated using examples with known labels. Several biologically-inspired classification tools are proposed and compared, including population-level confidence rating, and n-grams using spike motif algorithm. Using the optimal choice of parameters, our approach produces improvements over state-of-art spiking neural networks.
BindsNET: A machine learning-oriented spiking neural networks library in Python
Jun 04, 2018



Abstract:The development of spiking neural network simulation software is a critical component enabling the modeling of neural systems and the development of biologically inspired algorithms. Existing software frameworks support a wide range of neural functionality, software abstraction levels, and hardware devices, yet are typically not suitable for rapid prototyping or application to problems in the domain of machine learning. In this paper, we describe a new Python package for the simulation of spiking neural networks, specifically geared towards machine learning and reinforcement learning. Our software, called BindsNET, enables rapid building and simulation of spiking networks and features user-friendly, concise syntax. BindsNET is built on top of the PyTorch deep neural networks library, enabling fast CPU and GPU computation for large spiking networks. The BindsNET framework can be adjusted to meet the needs of other existing computing and hardware environments, e.g., TensorFlow. We also provide an interface into the OpenAI gym library, allowing for training and evaluation of spiking networks on reinforcement learning problems. We argue that this package facilitates the use of spiking networks for large-scale machine learning experimentation, and show some simple examples of how we envision BindsNET can be used in practice. BindsNET code is available at https://github.com/Hananel-Hazan/bindsnet
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge