Darpan T. Sanghavi
Lattice Map Spiking Neural Networks (LM-SNNs) for Clustering and Classifying Image Data
Jun 04, 2019
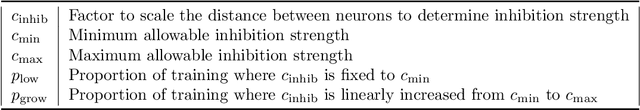
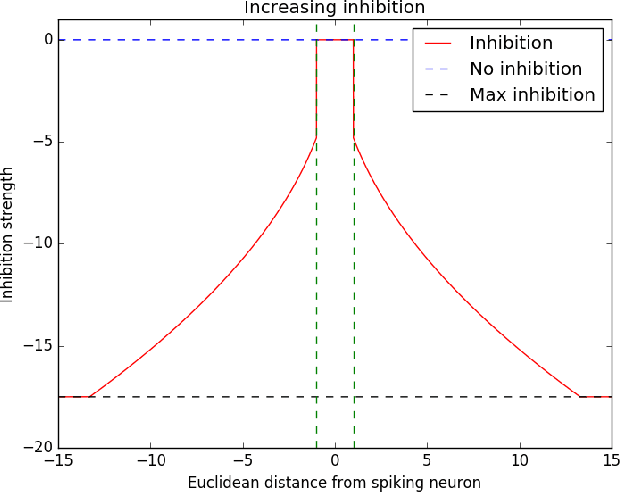
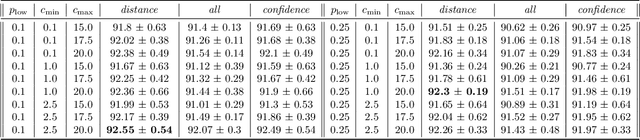
Abstract:Spiking neural networks (SNNs) with a lattice architecture are introduced in this work, combining several desirable properties of SNNs and self-organized maps (SOMs). Networks are trained with biologically motivated, unsupervised learning rules to obtain a self-organized grid of filters via cooperative and competitive excitatory-inhibitory interactions. Several inhibition strategies are developed and tested, such as (i) incrementally increasing inhibition level over the course of network training, and (ii) switching the inhibition level from low to high (two-level) after an initial training segment. During the labeling phase, the spiking activity generated by data with known labels is used to assign neurons to categories of data, which are then used to evaluate the network's classification ability on a held-out set of test data. Several biologically plausible evaluation rules are proposed and compared, including a population-level confidence rating, and an $n$-gram inspired method. The effectiveness of the proposed self-organized learning mechanism is tested using the MNIST benchmark dataset, as well as using images produced by playing the Atari Breakout game.
Unsupervised Learning with Self-Organizing Spiking Neural Networks
Jul 24, 2018



Abstract:We present a system comprising a hybridization of self-organized map (SOM) properties with spiking neural networks (SNNs) that retain many of the features of SOMs. Networks are trained in an unsupervised manner to learn a self-organized lattice of filters via excitatory-inhibitory interactions among populations of neurons. We develop and test various inhibition strategies, such as growing with inter-neuron distance and two distinct levels of inhibition. The quality of the unsupervised learning algorithm is evaluated using examples with known labels. Several biologically-inspired classification tools are proposed and compared, including population-level confidence rating, and n-grams using spike motif algorithm. Using the optimal choice of parameters, our approach produces improvements over state-of-art spiking neural networks.
BindsNET: A machine learning-oriented spiking neural networks library in Python
Jun 04, 2018



Abstract:The development of spiking neural network simulation software is a critical component enabling the modeling of neural systems and the development of biologically inspired algorithms. Existing software frameworks support a wide range of neural functionality, software abstraction levels, and hardware devices, yet are typically not suitable for rapid prototyping or application to problems in the domain of machine learning. In this paper, we describe a new Python package for the simulation of spiking neural networks, specifically geared towards machine learning and reinforcement learning. Our software, called BindsNET, enables rapid building and simulation of spiking networks and features user-friendly, concise syntax. BindsNET is built on top of the PyTorch deep neural networks library, enabling fast CPU and GPU computation for large spiking networks. The BindsNET framework can be adjusted to meet the needs of other existing computing and hardware environments, e.g., TensorFlow. We also provide an interface into the OpenAI gym library, allowing for training and evaluation of spiking networks on reinforcement learning problems. We argue that this package facilitates the use of spiking networks for large-scale machine learning experimentation, and show some simple examples of how we envision BindsNET can be used in practice. BindsNET code is available at https://github.com/Hananel-Hazan/bindsnet
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge