Cooper Sigrist
Improving Recommendation System Serendipity Through Lexicase Selection
May 18, 2023
Abstract:Recommender systems influence almost every aspect of our digital lives. Unfortunately, in striving to give us what we want, they end up restricting our open-mindedness. Current recommender systems promote echo chambers, where people only see the information they want to see, and homophily, where users of similar background see similar content. We propose a new serendipity metric to measure the presence of echo chambers and homophily in recommendation systems using cluster analysis. We then attempt to improve the diversity-preservation qualities of well known recommendation techniques by adopting a parent selection algorithm from the evolutionary computation literature known as lexicase selection. Our results show that lexicase selection, or a mixture of lexicase selection and ranking, outperforms its purely ranked counterparts in terms of personalization, coverage and our specifically designed serendipity benchmark, while only slightly under-performing in terms of accuracy (hit rate). We verify these results across a variety of recommendation list sizes. In this work we show that lexicase selection is able to maintain multiple diverse clusters of item recommendations that are each relevant for the specific user, while still maintaining a high hit-rate accuracy, a trade off that is not achieved by other methods.
Minibatch Processing in Spiking Neural Networks
Sep 05, 2019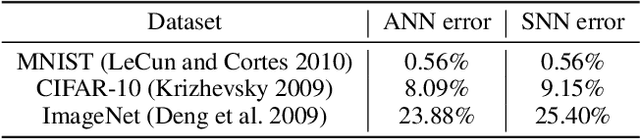
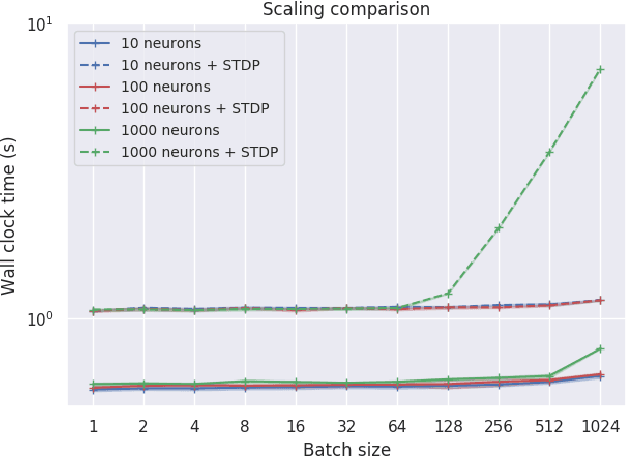

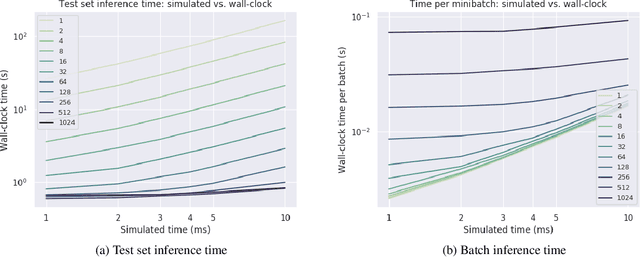
Abstract:Spiking neural networks (SNNs) are a promising candidate for biologically-inspired and energy efficient computation. However, their simulation is notoriously time consuming, and may be seen as a bottleneck in developing competitive training methods with potential deployment on neuromorphic hardware platforms. To address this issue, we provide an implementation of mini-batch processing applied to clock-based SNN simulation, leading to drastically increased data throughput. To our knowledge, this is the first general-purpose implementation of mini-batch processing in a spiking neural networks simulator, which works with arbitrary neuron and synapse models. We demonstrate nearly constant-time scaling with batch size on a simulation setup (up to GPU memory limits), and showcase the effectiveness of large batch sizes in two SNN application domains, resulting in $\approx$880X and $\approx$24X reductions in wall-clock time respectively. Different parameter reduction techniques are shown to produce different learning outcomes in a simulation of networks trained with spike-timing-dependent plasticity. Machine learning practitioners and biological modelers alike may benefit from the drastically reduced simulation time and increased iteration speed this method enables. Code to reproduce the benchmarks and experimental findings in this paper can be found at https://github.com/djsaunde/snn-minibatch.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge