Cristina L. Saratxaga
Unleashing the Potential of Synthetic Images: A Study on Histopathology Image Classification
Sep 24, 2024Abstract:Histopathology image classification is crucial for the accurate identification and diagnosis of various diseases but requires large and diverse datasets. Obtaining such datasets, however, is often costly and time-consuming due to the need for expert annotations and ethical constraints. To address this, we examine the suitability of different generative models and image selection approaches to create realistic synthetic histopathology image patches conditioned on class labels. Our findings highlight the importance of selecting an appropriate generative model type and architecture to enhance performance. Our experiments over the PCam dataset show that diffusion models are effective for transfer learning, while GAN-generated samples are better suited for augmentation. Additionally, transformer-based generative models do not require image filtering, in contrast to those derived from Convolutional Neural Networks (CNNs), which benefit from realism score-based selection. Therefore, we show that synthetic images can effectively augment existing datasets, ultimately improving the performance of the downstream histopathology image classification task.
Data-Driven Color Augmentation Techniques for Deep Skin Image Analysis
Mar 10, 2017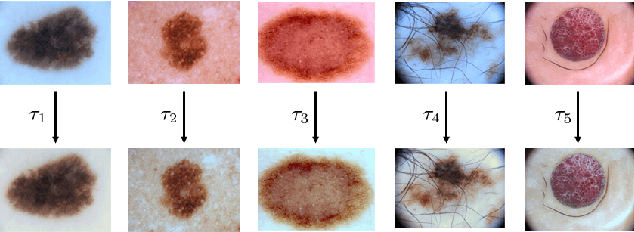
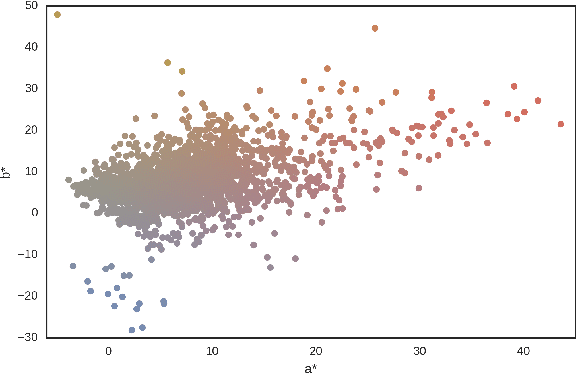
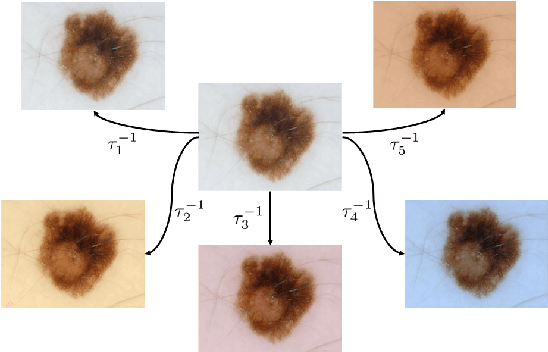
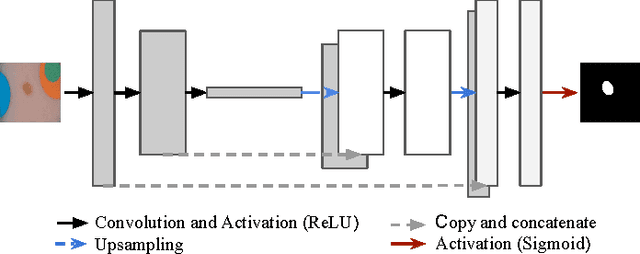
Abstract:Dermoscopic skin images are often obtained with different imaging devices, under varying acquisition conditions. In this work, instead of attempting to perform intensity and color normalization, we propose to leverage computational color constancy techniques to build an artificial data augmentation technique suitable for this kind of images. Specifically, we apply the \emph{shades of gray} color constancy technique to color-normalize the entire training set of images, while retaining the estimated illuminants. We then draw one sample from the distribution of training set illuminants and apply it on the normalized image. We employ this technique for training two deep convolutional neural networks for the tasks of skin lesion segmentation and skin lesion classification, in the context of the ISIC 2017 challenge and without using any external dermatologic image set. Our results on the validation set are promising, and will be supplemented with extended results on the hidden test set when available.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge