Christy Li
Automated Detection of Visual Attribute Reliance with a Self-Reflective Agent
Oct 24, 2025Abstract:When a vision model performs image recognition, which visual attributes drive its predictions? Detecting unintended reliance on specific visual features is critical for ensuring model robustness, preventing overfitting, and avoiding spurious correlations. We introduce an automated framework for detecting such dependencies in trained vision models. At the core of our method is a self-reflective agent that systematically generates and tests hypotheses about visual attributes that a model may rely on. This process is iterative: the agent refines its hypotheses based on experimental outcomes and uses a self-evaluation protocol to assess whether its findings accurately explain model behavior. When inconsistencies arise, the agent self-reflects over its findings and triggers a new cycle of experimentation. We evaluate our approach on a novel benchmark of 130 models designed to exhibit diverse visual attribute dependencies across 18 categories. Our results show that the agent's performance consistently improves with self-reflection, with a significant performance increase over non-reflective baselines. We further demonstrate that the agent identifies real-world visual attribute dependencies in state-of-the-art models, including CLIP's vision encoder and the YOLOv8 object detector.
Convolutional Neural Networks for Medical Diagnosis from Admission Notes
Dec 06, 2017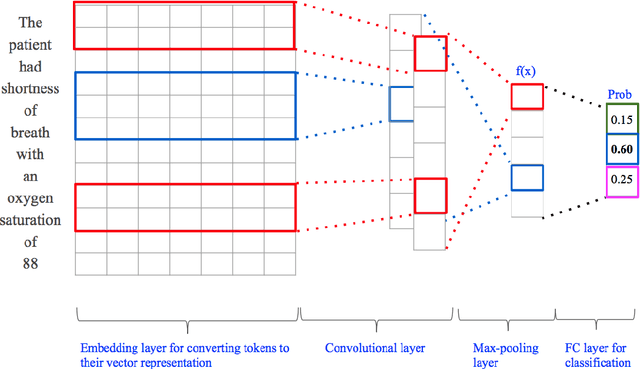
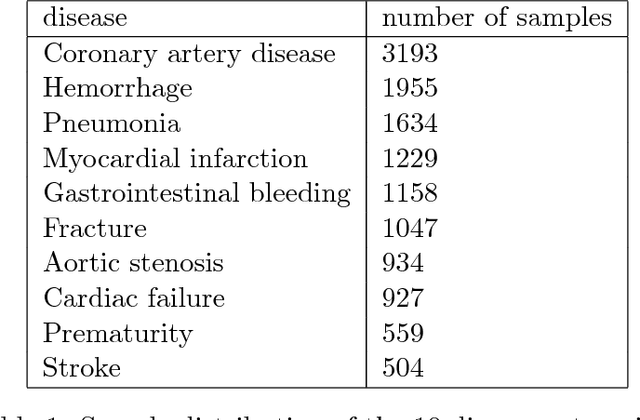
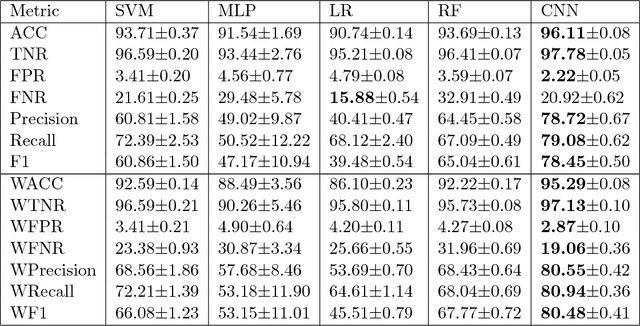
Abstract:$\textbf{Objective}$ Develop an automatic diagnostic system which only uses textual admission information from Electronic Health Records (EHRs) and assist clinicians with a timely and statistically proved decision tool. The hope is that the tool can be used to reduce mis-diagnosis. $\textbf{Materials and Methods}$ We use the real-world clinical notes from MIMIC-III, a freely available dataset consisting of clinical data of more than forty thousand patients who stayed in intensive care units of the Beth Israel Deaconess Medical Center between 2001 and 2012. We proposed a Convolutional Neural Network model to learn semantic features from unstructured textual input and automatically predict primary discharge diagnosis. $\textbf{Results}$ The proposed model achieved an overall 96.11% accuracy and 80.48% weighted F1 score values on 10 most frequent disease classes, significantly outperforming four strong baseline models by at least 12.7% in weighted F1 score. $\textbf{Discussion}$ Experimental results imply that the CNN model is suitable for supporting diagnosis decision making in the presence of complex, noisy and unstructured clinical data while at the same time using fewer layers and parameters that other traditional Deep Network models. $\textbf{Conclusion}$ Our model demonstrated capability of representing complex medical meaningful features from unstructured clinical notes and prediction power for commonly misdiagnosed frequent diseases. It can use easily adopted in clinical setting to provide timely and statistically proved decision support. $\textbf{Keywords}$ Convolutional neural network, text classification, discharge diagnosis prediction, admission information from EHRs.
Predicting Discharge Medications at Admission Time Based on Deep Learning
Dec 05, 2017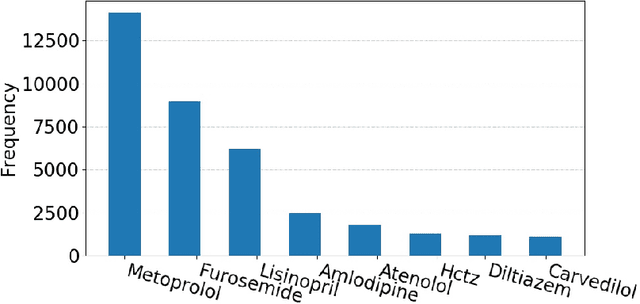

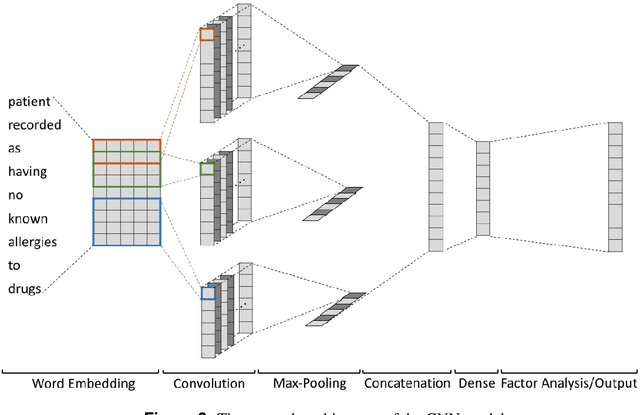
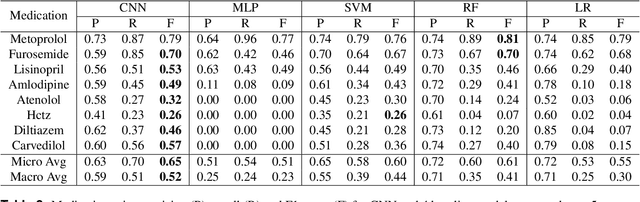
Abstract:Predicting discharge medications right after a patient being admitted is an important clinical decision, which provides physicians with guidance on what type of medication regimen to plan for and what possible changes on initial medication may occur during an inpatient stay. It also facilitates medication reconciliation process with easy detection of medication discrepancy at discharge time to improve patient safety. However, since the information available upon admission is limited and patients' condition may evolve during an inpatient stay, these predictions could be a difficult decision for physicians to make. In this work, we investigate how to leverage deep learning technologies to assist physicians in predicting discharge medications based on information documented in the admission note. We build a convolutional neural network which takes an admission note as input and predicts the medications placed on the patient at discharge time. Our method is able to distill semantic patterns from unstructured and noisy texts, and is capable of capturing the pharmacological correlations among medications. We evaluate our method on 25K patient visits and compare with 4 strong baselines. Our methods demonstrate a 20% increase in macro-averaged F1 score than the best baseline.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge