Christopher R. Collins
Tuning Hyperparameters without Grad Students: Scalable and Robust Bayesian Optimisation with Dragonfly
Mar 15, 2019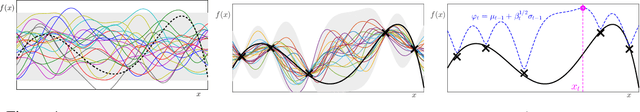
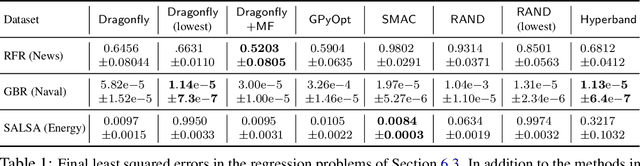
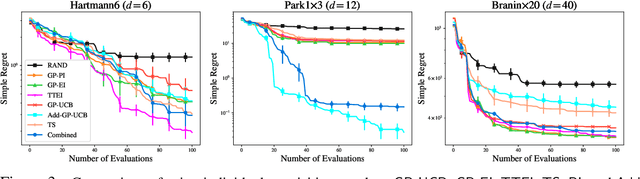
Abstract:Bayesian Optimisation (BO), refers to a suite of techniques for global optimisation of expensive black box functions, which use introspective Bayesian models of the function to efficiently find the optimum. While BO has been applied successfully in many applications, modern optimisation tasks usher in new challenges where conventional methods fail spectacularly. In this work, we present Dragonfly, an open source Python library for scalable and robust BO. Dragonfly incorporates multiple recently developed methods that allow BO to be applied in challenging real world settings; these include better methods for handling higher dimensional domains, methods for handling multi-fidelity evaluations when cheap approximations of an expensive function are available, methods for optimising over structured combinatorial spaces, such as the space of neural network architectures, and methods for handling parallel evaluations. Additionally, we develop new methodological improvements in BO for selecting the Bayesian model, selecting the acquisition function, and optimising over complex domains with different variable types and additional constraints. We compare Dragonfly to a suite of other packages and algorithms for global optimisation and demonstrate that when the above methods are integrated, they enable significant improvements in the performance of BO. The Dragonfly library is available at dragonfly.github.io.
Constant Size Molecular Descriptors For Use With Machine Learning
Jan 23, 2017
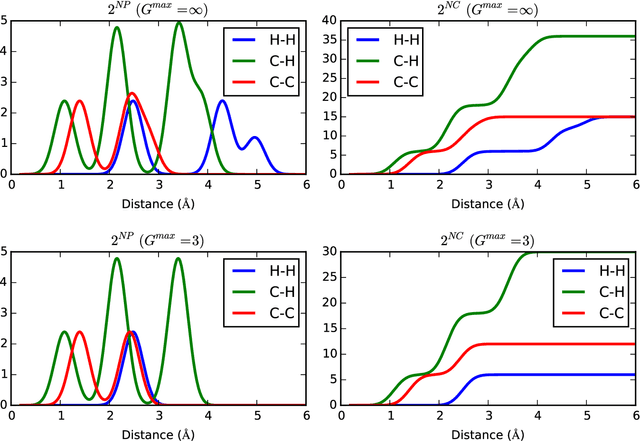
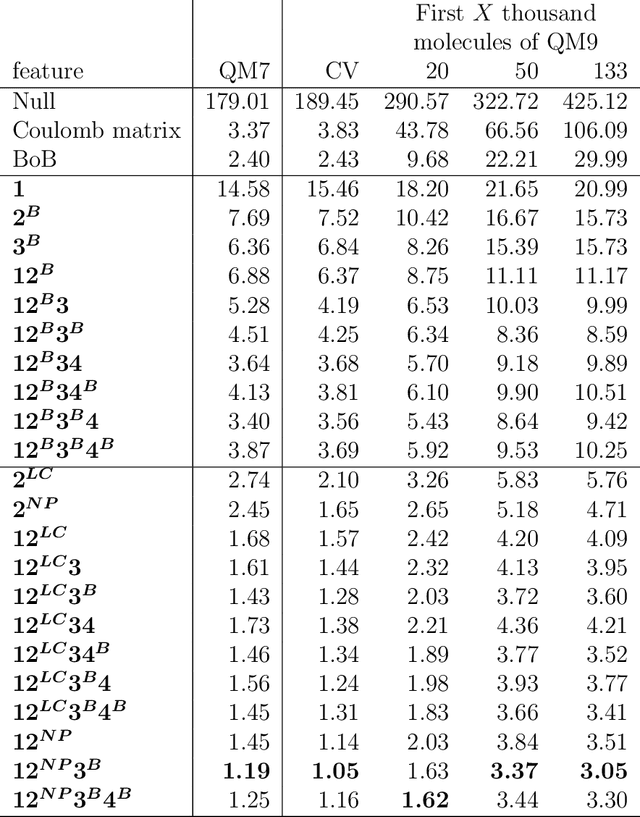
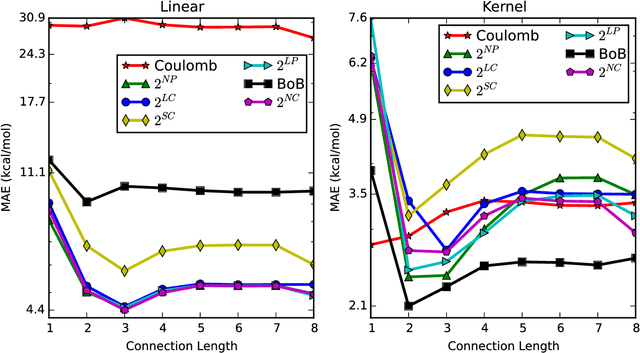
Abstract:A set of molecular descriptors whose length is independent of molecular size is developed for machine learning models that target thermodynamic and electronic properties of molecules. These features are evaluated by monitoring performance of kernel ridge regression models on well-studied data sets of small organic molecules. The features include connectivity counts, which require only the bonding pattern of the molecule, and encoded distances, which summarize distances between both bonded and non-bonded atoms and so require the full molecular geometry. In addition to having constant size, these features summarize information regarding the local environment of atoms and bonds, such that models can take advantage of similarities resulting from the presence of similar chemical fragments across molecules. Combining these two types of features leads to models whose performance is comparable to or better than the current state of the art. The features introduced here have the advantage of leading to models that may be trained on smaller molecules and then used successfully on larger molecules.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge