Christopher Chute
A Methodological Framework for the Comparative Evaluation of Multiple Imputation Methods: Multiple Imputation of Race, Ethnicity and Body Mass Index in the U.S. National COVID Cohort Collaborative
Jun 13, 2022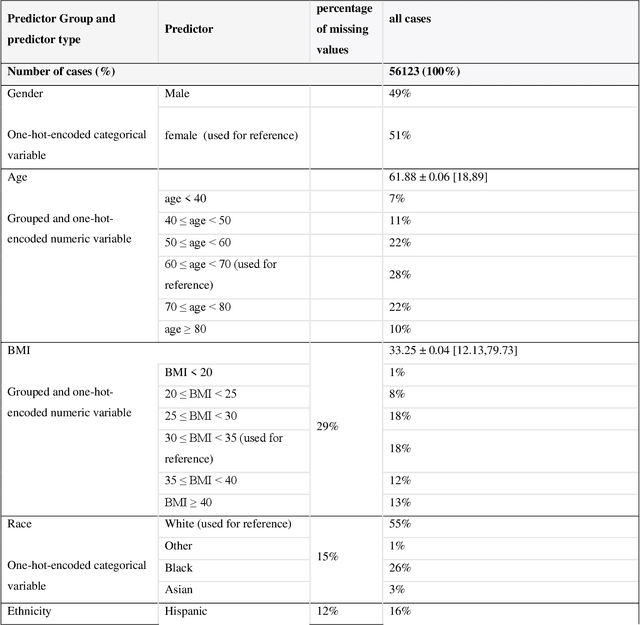
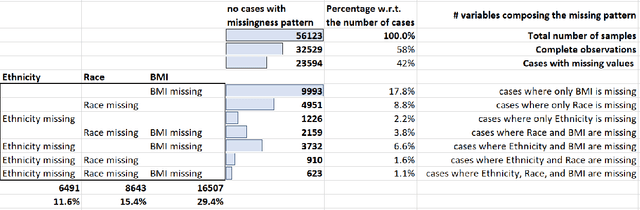
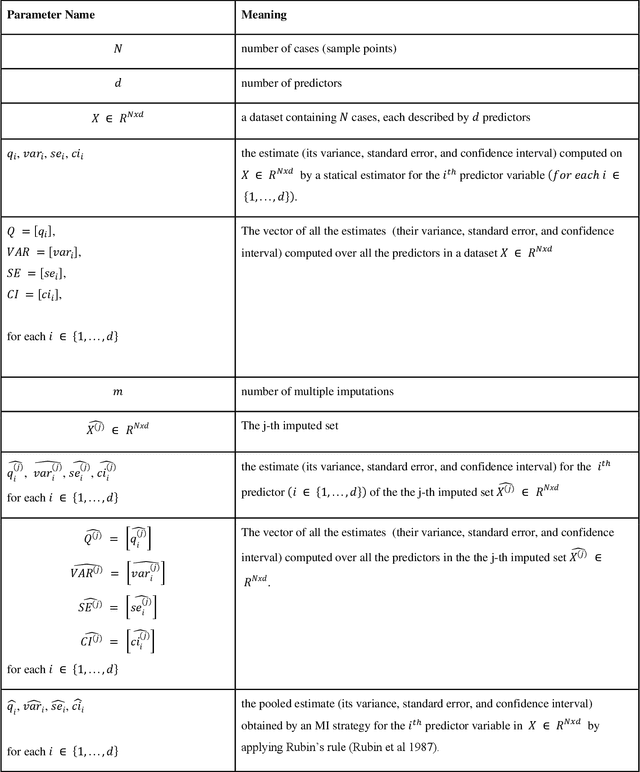
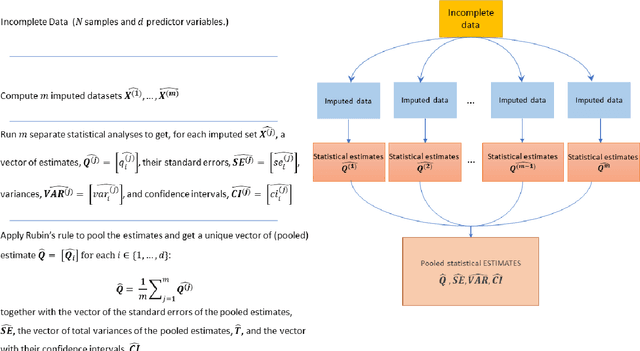
Abstract:While electronic health records are a rich data source for biomedical research, these systems are not implemented uniformly across healthcare settings and significant data may be missing due to healthcare fragmentation and lack of interoperability between siloed electronic health records. Considering that the deletion of cases with missing data may introduce severe bias in the subsequent analysis, several authors prefer applying a multiple imputation strategy to recover the missing information. Unfortunately, although several literature works have documented promising results by using any of the different multiple imputation algorithms that are now freely available for research, there is no consensus on which MI algorithm works best. Beside the choice of the MI strategy, the choice of the imputation algorithm and its application settings are also both crucial and challenging. In this paper, inspired by the seminal works of Rubin and van Buuren, we propose a methodological framework that may be applied to evaluate and compare several multiple imputation techniques, with the aim to choose the most valid for computing inferences in a clinical research work. Our framework has been applied to validate, and extend on a larger cohort, the results we presented in a previous literature study, where we evaluated the influence of crucial patients' descriptors and COVID-19 severity in patients with type 2 diabetes mellitus whose data is provided by the National COVID Cohort Collaborative Enclave.
AlignFlow: Cycle Consistent Learning from Multiple Domains via Normalizing Flows
May 30, 2019



Abstract:Given unpaired data from multiple domains, a key challenge is to efficiently exploit these data sources for modeling a target domain. Variants of this problem have been studied in many contexts, such as cross-domain translation and domain adaptation. We propose AlignFlow, a generative modeling framework for learning from multiple domains via normalizing flows. The use of normalizing flows in AlignFlow allows for a) flexibility in specifying learning objectives via adversarial training, maximum likelihood estimation, or a hybrid of the two methods; and b) exact inference of the shared latent factors across domains at test time. We derive theoretical results for the conditions under which AlignFlow guarantees marginal consistency for the different learning objectives. Furthermore, we show that AlignFlow guarantees exact cycle consistency in mapping datapoints from one domain to another. Empirically, AlignFlow can be used for data-efficient density estimation given multiple data sources and shows significant improvements over relevant baselines on unsupervised domain adaptation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge