Christian Widmer
End-to-End Annotator Bias Approximation on Crowdsourced Single-Label Sentiment Analysis
Nov 03, 2021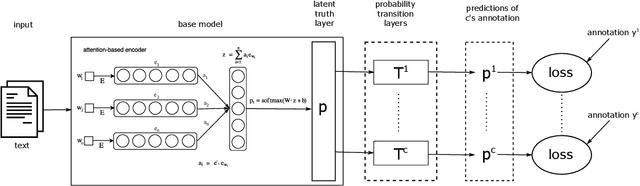
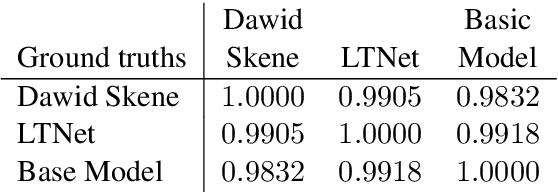
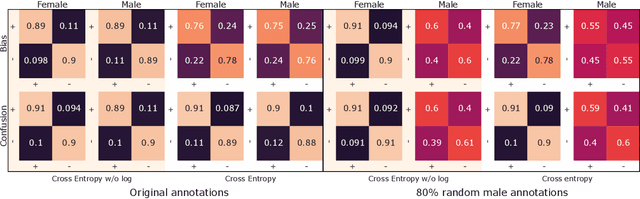
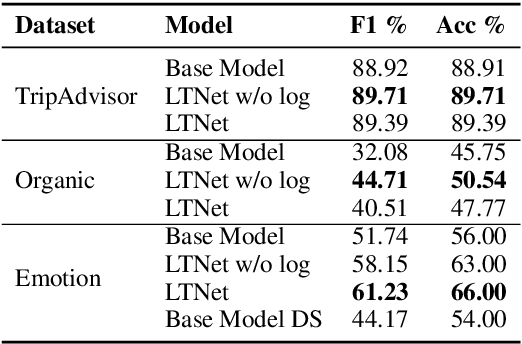
Abstract:Sentiment analysis is often a crowdsourcing task prone to subjective labels given by many annotators. It is not yet fully understood how the annotation bias of each annotator can be modeled correctly with state-of-the-art methods. However, resolving annotator bias precisely and reliably is the key to understand annotators' labeling behavior and to successfully resolve corresponding individual misconceptions and wrongdoings regarding the annotation task. Our contribution is an explanation and improvement for precise neural end-to-end bias modeling and ground truth estimation, which reduces an undesired mismatch in that regard of the existing state-of-the-art. Classification experiments show that it has potential to improve accuracy in cases where each sample is annotated only by one single annotator. We provide the whole source code publicly and release an own domain-specific sentiment dataset containing 10,000 sentences discussing organic food products. These are crawled from social media and are singly labeled by 10 non-expert annotators.
* 10 pages, 2 figures, 2 tables, full conference paper, peer-reviewed
Framework for Multi-task Multiple Kernel Learning and Applications in Genome Analysis
Jun 30, 2015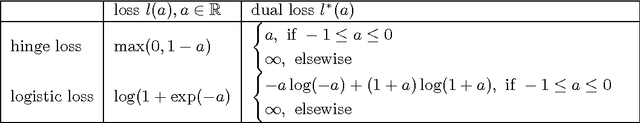
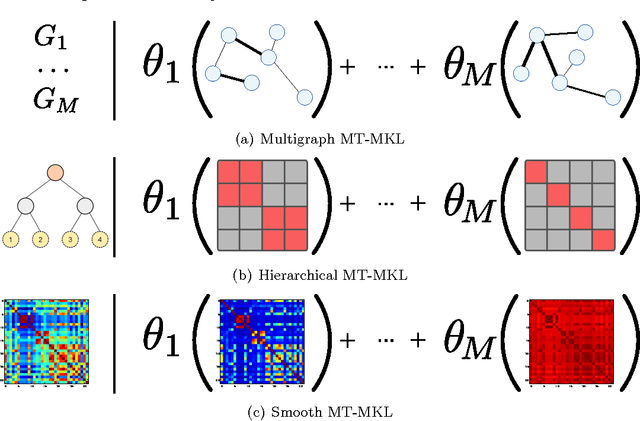
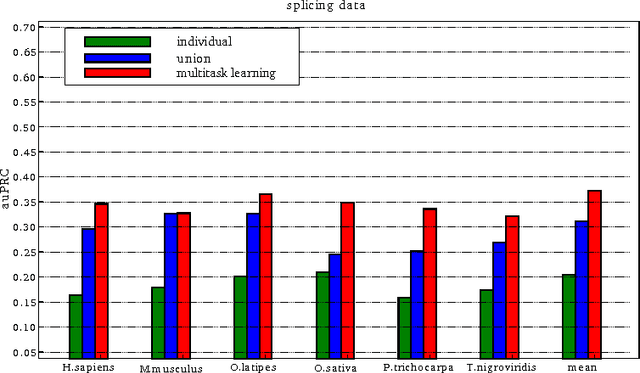
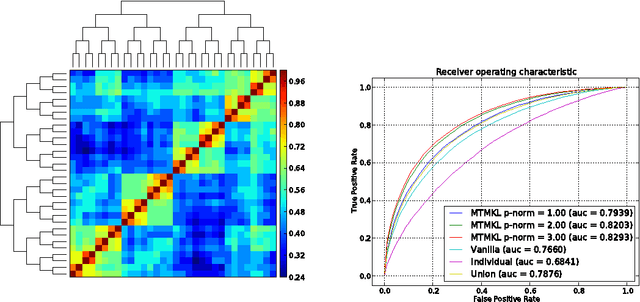
Abstract:We present a general regularization-based framework for Multi-task learning (MTL), in which the similarity between tasks can be learned or refined using $\ell_p$-norm Multiple Kernel learning (MKL). Based on this very general formulation (including a general loss function), we derive the corresponding dual formulation using Fenchel duality applied to Hermitian matrices. We show that numerous established MTL methods can be derived as special cases from both, the primal and dual of our formulation. Furthermore, we derive a modern dual-coordinate descend optimization strategy for the hinge-loss variant of our formulation and provide convergence bounds for our algorithm. As a special case, we implement in C++ a fast LibLinear-style solver for $\ell_p$-norm MKL. In the experimental section, we analyze various aspects of our algorithm such as predictive performance and ability to reconstruct task relationships on biologically inspired synthetic data, where we have full control over the underlying ground truth. We also experiment on a new dataset from the domain of computational biology that we collected for the purpose of this paper. It concerns the prediction of transcription start sites (TSS) over nine organisms, which is a crucial task in gene finding. Our solvers including all discussed special cases are made available as open-source software as part of the SHOGUN machine learning toolbox (available at \url{http://shogun.ml}).
GRED: Graph-Regularized 3D Shape Reconstruction from Highly Anisotropic and Noisy Images
Sep 17, 2013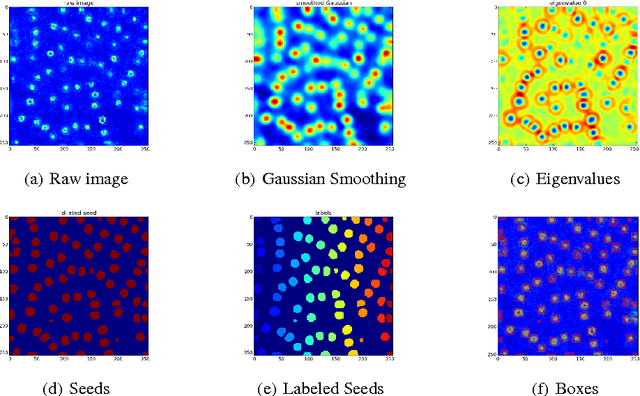

Abstract:Analysis of microscopy images can provide insight into many biological processes. One particularly challenging problem is cell nuclear segmentation in highly anisotropic and noisy 3D image data. Manually localizing and segmenting each and every cell nuclei is very time consuming, which remains a bottleneck in large scale biological experiments. In this work we present a tool for automated segmentation of cell nuclei from 3D fluorescent microscopic data. Our tool is based on state-of-the-art image processing and machine learning techniques and supports a friendly graphical user interface (GUI). We show that our tool is as accurate as manual annotation but greatly reduces the time for the registration.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge