Chloe O'Connell
Impact of Large Language Model Assistance on Patients Reading Clinical Notes: A Mixed-Methods Study
Jan 17, 2024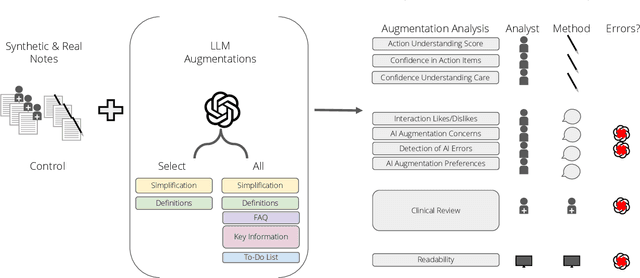
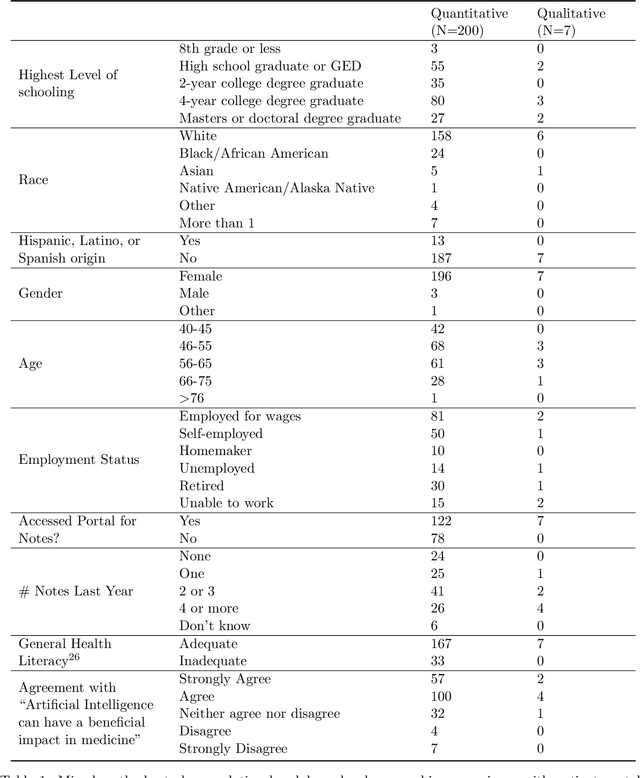
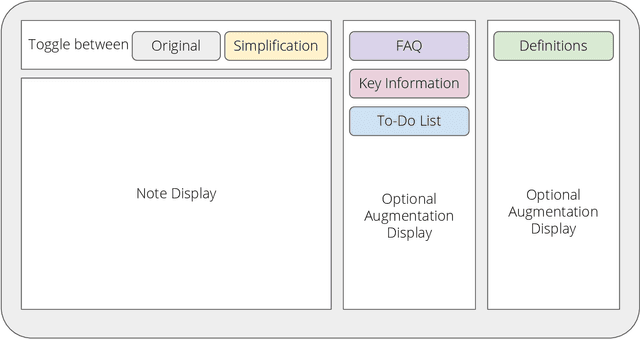
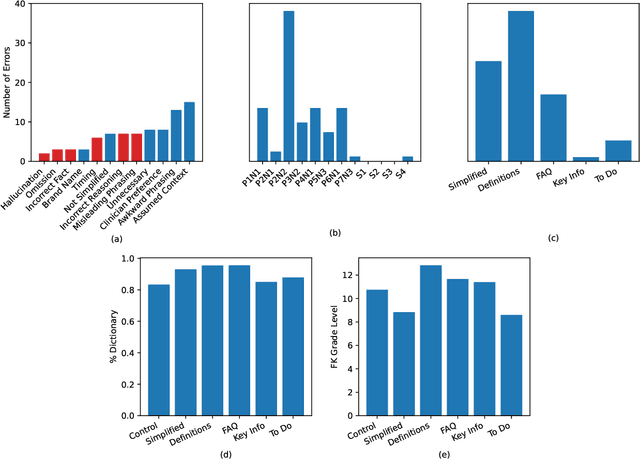
Abstract:Patients derive numerous benefits from reading their clinical notes, including an increased sense of control over their health and improved understanding of their care plan. However, complex medical concepts and jargon within clinical notes hinder patient comprehension and may lead to anxiety. We developed a patient-facing tool to make clinical notes more readable, leveraging large language models (LLMs) to simplify, extract information from, and add context to notes. We prompt engineered GPT-4 to perform these augmentation tasks on real clinical notes donated by breast cancer survivors and synthetic notes generated by a clinician, a total of 12 notes with 3868 words. In June 2023, 200 female-identifying US-based participants were randomly assigned three clinical notes with varying levels of augmentations using our tool. Participants answered questions about each note, evaluating their understanding of follow-up actions and self-reported confidence. We found that augmentations were associated with a significant increase in action understanding score (0.63 $\pm$ 0.04 for select augmentations, compared to 0.54 $\pm$ 0.02 for the control) with p=0.002. In-depth interviews of self-identifying breast cancer patients (N=7) were also conducted via video conferencing. Augmentations, especially definitions, elicited positive responses among the seven participants, with some concerns about relying on LLMs. Augmentations were evaluated for errors by clinicians, and we found misleading errors occur, with errors more common in real donated notes than synthetic notes, illustrating the importance of carefully written clinical notes. Augmentations improve some but not all readability metrics. This work demonstrates the potential of LLMs to improve patients' experience with clinical notes at a lower burden to clinicians. However, having a human in the loop is important to correct potential model errors.
Robust Benchmarking for Machine Learning of Clinical Entity Extraction
Jul 31, 2020
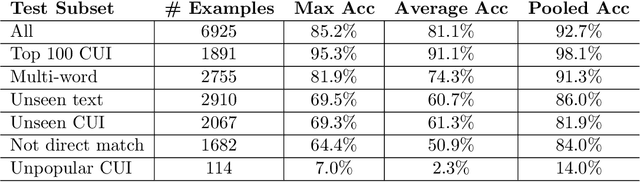
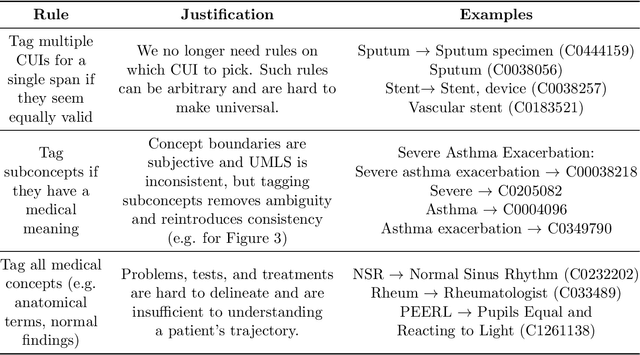
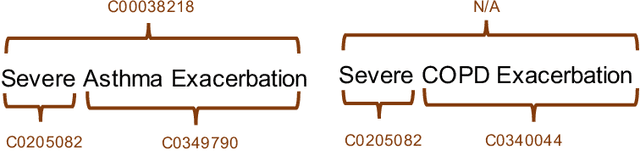
Abstract:Clinical studies often require understanding elements of a patient's narrative that exist only in free text clinical notes. To transform notes into structured data for downstream use, these elements are commonly extracted and normalized to medical vocabularies. In this work, we audit the performance of and indicate areas of improvement for state-of-the-art systems. We find that high task accuracies for clinical entity normalization systems on the 2019 n2c2 Shared Task are misleading, and underlying performance is still brittle. Normalization accuracy is high for common concepts (95.3%), but much lower for concepts unseen in training data (69.3%). We demonstrate that current approaches are hindered in part by inconsistencies in medical vocabularies, limitations of existing labeling schemas, and narrow evaluation techniques. We reformulate the annotation framework for clinical entity extraction to factor in these issues to allow for robust end-to-end system benchmarking. We evaluate concordance of annotations from our new framework between two annotators and achieve a Jaccard similarity of 0.73 for entity recognition and an agreement of 0.83 for entity normalization. We propose a path forward to address the demonstrated need for the creation of a reference standard to spur method development in entity recognition and normalization.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge