Robust Benchmarking for Machine Learning of Clinical Entity Extraction
Paper and Code
Jul 31, 2020
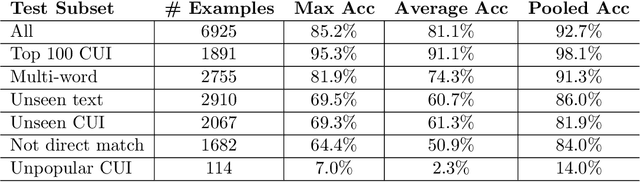
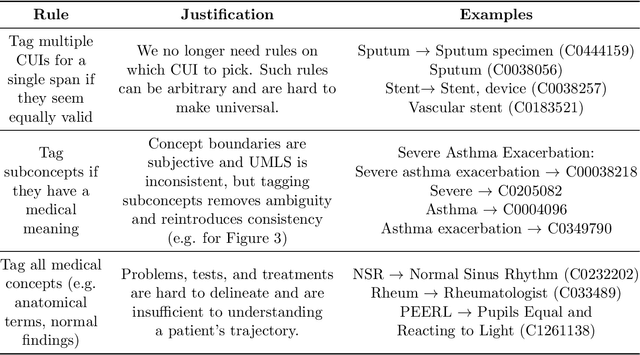
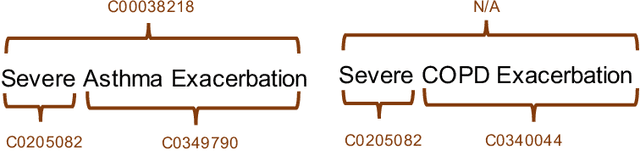
Clinical studies often require understanding elements of a patient's narrative that exist only in free text clinical notes. To transform notes into structured data for downstream use, these elements are commonly extracted and normalized to medical vocabularies. In this work, we audit the performance of and indicate areas of improvement for state-of-the-art systems. We find that high task accuracies for clinical entity normalization systems on the 2019 n2c2 Shared Task are misleading, and underlying performance is still brittle. Normalization accuracy is high for common concepts (95.3%), but much lower for concepts unseen in training data (69.3%). We demonstrate that current approaches are hindered in part by inconsistencies in medical vocabularies, limitations of existing labeling schemas, and narrow evaluation techniques. We reformulate the annotation framework for clinical entity extraction to factor in these issues to allow for robust end-to-end system benchmarking. We evaluate concordance of annotations from our new framework between two annotators and achieve a Jaccard similarity of 0.73 for entity recognition and an agreement of 0.83 for entity normalization. We propose a path forward to address the demonstrated need for the creation of a reference standard to spur method development in entity recognition and normalization.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge