Catherine Lebel
Cumming School of Medicine, Department of Radiology, University of Calgary, Calgary, Canada
Personalized White Matter Bundle Segmentation for Early Childhood
Feb 04, 2026Abstract:White matter segmentation methods from diffusion magnetic resonance imaging range from streamline clustering-based approaches to bundle mask delineation, but none have proposed a pediatric-specific approach. We hypothesize that a deep learning model with a similar approach to TractSeg will improve similarity between an algorithm-generated mask and an expert-labeled ground truth. Given a cohort of 56 manually labelled white matter bundles, we take inspiration from TractSeg's 2D UNet architecture, and we modify inputs to match bundle definitions as determined by pediatric experts, evaluation to use k fold cross validation, the loss function to masked Dice loss. We evaluate Dice score, volume overlap, and volume overreach of 16 major regions of interest compared to the expert labeled dataset. To test whether our approach offers statistically significant improvements over TractSeg, we compare Dice voxels, volume overlap, and adjacency voxels with a Wilcoxon signed rank test followed by false discovery rate correction. We find statistical significance across all bundles for all metrics with one exception in volume overlap. After we run TractSeg and our model, we combine their output masks into a 60 label atlas to evaluate if TractSeg and our model combined can generate a robust, individualized atlas, and observe smoothed, continuous masks in cases that TractSeg did not produce an anatomically plausible output. With the improvement of white matter pathway segmentation masks, we can further understand neurodevelopment on a population level scale, and we can produce reliable estimates of individualized anatomy in pediatric white matter diseases and disorders.
Trustworthy and Responsible AI for Human-Centric Autonomous Decision-Making Systems
Sep 02, 2024Abstract:Artificial Intelligence (AI) has paved the way for revolutionary decision-making processes, which if harnessed appropriately, can contribute to advancements in various sectors, from healthcare to economics. However, its black box nature presents significant ethical challenges related to bias and transparency. AI applications are hugely impacted by biases, presenting inconsistent and unreliable findings, leading to significant costs and consequences, highlighting and perpetuating inequalities and unequal access to resources. Hence, developing safe, reliable, ethical, and Trustworthy AI systems is essential. Our team of researchers working with Trustworthy and Responsible AI, part of the Transdisciplinary Scholarship Initiative within the University of Calgary, conducts research on Trustworthy and Responsible AI, including fairness, bias mitigation, reproducibility, generalization, interpretability, and authenticity. In this paper, we review and discuss the intricacies of AI biases, definitions, methods of detection and mitigation, and metrics for evaluating bias. We also discuss open challenges with regard to the trustworthiness and widespread application of AI across diverse domains of human-centric decision making, as well as guidelines to foster Responsible and Trustworthy AI models.
Generalizing Deep Whole Brain Segmentation for Pediatric and Post-Contrast MRI with Augmented Transfer Learning
Aug 13, 2019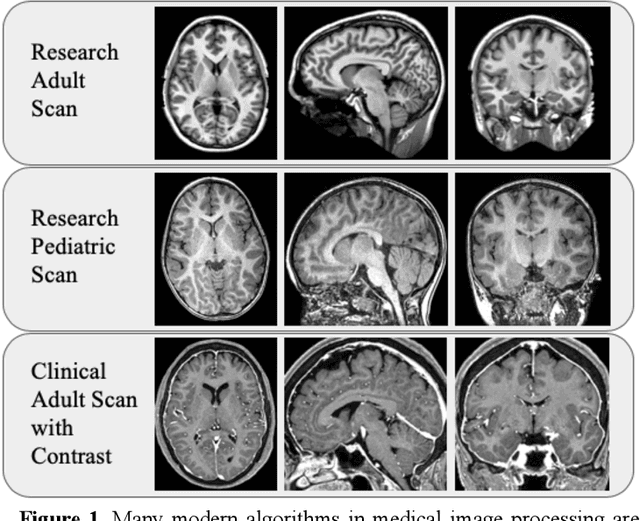
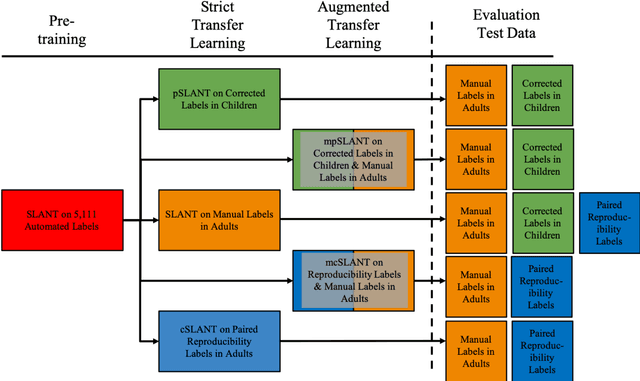

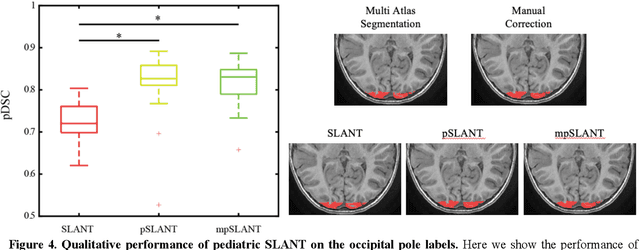
Abstract:Generalizability is an important problem in deep neural networks, especially in the context of the variability of data acquisition in clinical magnetic resonance imaging (MRI). Recently, the Spatially Localized Atlas Network Tiles (SLANT) approach has been shown to effectively segment whole brain non-contrast T1w MRI with 132 volumetric labels. Enhancing generalizability of SLANT would enable broader application of volumetric assessment in multi-site studies. Transfer learning (TL) is commonly used to update the neural network weights for local factors; yet, it is commonly recognized to risk degradation of performance on the original validation/test cohorts. Here, we explore TL by data augmentation to address these concerns in the context of adapting SLANT to anatomical variation and scanning protocol. We consider two datasets: First, we optimize for age with 30 T1w MRI of young children with manually corrected volumetric labels, and accuracy of automated segmentation defined relative to the manually provided truth. Second, we optimize for acquisition with 36 paired datasets of pre- and post-contrast clinically acquired T1w MRI, and accuracy of the post-contrast segmentations assessed relative to the pre-contrast automated assessment. For both studies, we augment the original TL step of SLANT with either only the new data or with both original and new data. Over baseline SLANT, both approaches yielded significantly improved performance (signed rank tests; pediatric: 0.89 vs. 0.82 DSC, p<0.001; contrast: 0.80 vs 0.76, p<0.001). The performance on the original test set decreased with the new-data only transfer learning approach, so data augmentation was superior to strict transfer learning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge