Bernal Jimenez Gutierrez
Solving the Right Problem is Key for Translational NLP: A Case Study in UMLS Vocabulary Insertion
Nov 25, 2023
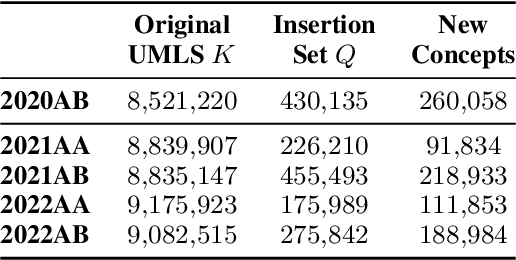


Abstract:As the immense opportunities enabled by large language models become more apparent, NLP systems will be increasingly expected to excel in real-world settings. However, in many instances, powerful models alone will not yield translational NLP solutions, especially if the formulated problem is not well aligned with the real-world task. In this work, we study the case of UMLS vocabulary insertion, an important real-world task in which hundreds of thousands of new terms, referred to as atoms, are added to the UMLS, one of the most comprehensive open-source biomedical knowledge bases. Previous work aimed to develop an automated NLP system to make this time-consuming, costly, and error-prone task more efficient. Nevertheless, practical progress in this direction has been difficult to achieve due to a problem formulation and evaluation gap between research output and the real-world task. In order to address this gap, we introduce a new formulation for UMLS vocabulary insertion which mirrors the real-world task, datasets which faithfully represent it and several strong baselines we developed through re-purposing existing solutions. Additionally, we propose an effective rule-enhanced biomedical language model which enables important new model behavior, outperforms all strong baselines and provides measurable qualitative improvements to editors who carry out the UVI task. We hope this case study provides insight into the considerable importance of problem formulation for the success of translational NLP solutions.
Clinical Reading Comprehension: A Thorough Analysis of the emrQA Dataset
May 01, 2020



Abstract:Machine reading comprehension has made great progress in recent years owing to large-scale annotated datasets. In the clinical domain, however, creating such datasets is quite difficult due to the domain expertise required for annotation. Recently, Pampari et al. (EMNLP'18) tackled this issue by using expert-annotated question templates and existing i2b2 annotations to create emrQA, the first large-scale dataset for question answering (QA) based on clinical notes. In this paper, we provide an in-depth analysis of this dataset and the clinical reading comprehension (CliniRC) task. From our qualitative analysis, we find that (i) emrQA answers are often incomplete, and (ii) emrQA questions are often answerable without using domain knowledge. From our quantitative experiments, surprising results include that (iii) using a small sampled subset (5%-20%), we can obtain roughly equal performance compared to the model trained on the entire dataset, (iv) this performance is close to human expert's performance, and (v) BERT models do not beat the best performing base model. Following our analysis of the emrQA, we further explore two desired aspects of CliniRC systems: the ability to utilize clinical domain knowledge and to generalize to unseen questions and contexts. We argue that both should be considered when creating future datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge