Ben Covington
Caveats in Generating Medical Imaging Labels from Radiology Reports
May 06, 2019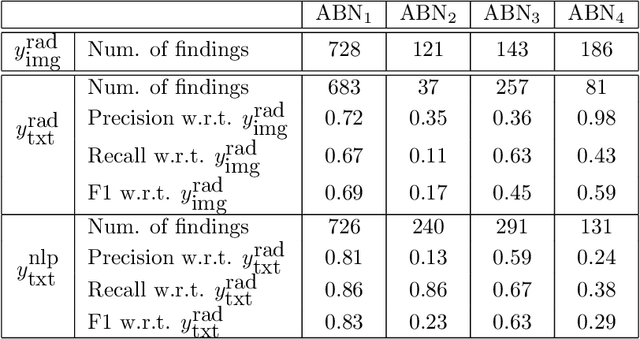
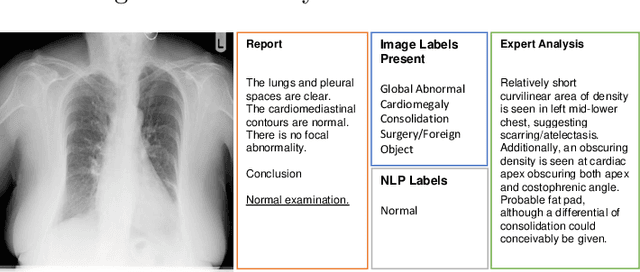
Abstract:Acquiring high-quality annotations in medical imaging is usually a costly process. Automatic label extraction with natural language processing (NLP) has emerged as a promising workaround to bypass the need of expert annotation. Despite the convenience, the limitation of such an approximation has not been carefully examined and is not well understood. With a challenging set of 1,000 chest X-ray studies and their corresponding radiology reports, we show that there exists a surprisingly large discrepancy between what radiologists visually perceive and what they clinically report. Furthermore, with inherently flawed report as ground truth, the state-of-the-art medical NLP fails to produce high-fidelity labels.
A Strong Baseline for Domain Adaptation and Generalization in Medical Imaging
Apr 02, 2019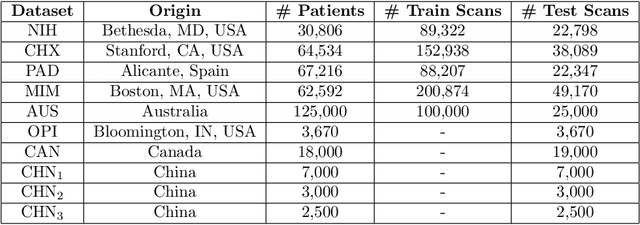
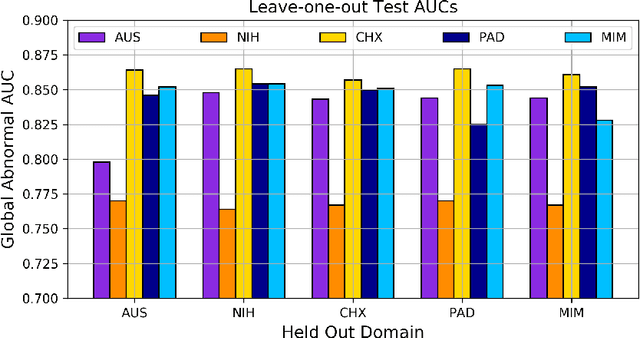
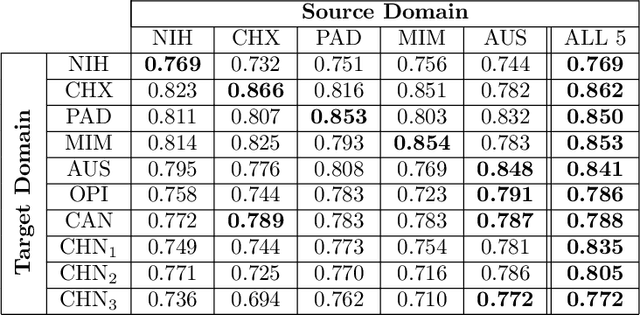
Abstract:This work provides a strong baseline for the problem of multi-source multi-target domain adaptation and generalization in medical imaging. Using a diverse collection of ten chest X-ray datasets, we empirically demonstrate the benefits of training medical imaging deep learning models on varied patient populations for generalization to out-of-sample domains.
Efficient and Accurate Abnormality Mining from Radiology Reports with Customized False Positive Reduction
Oct 01, 2018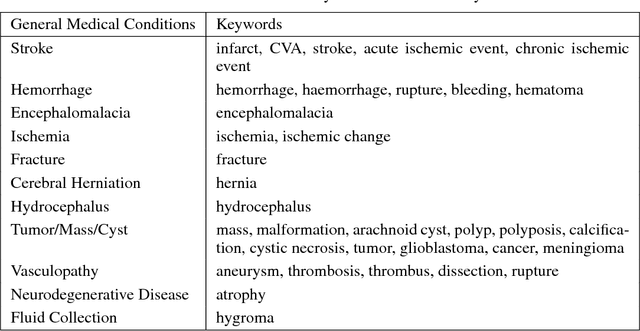
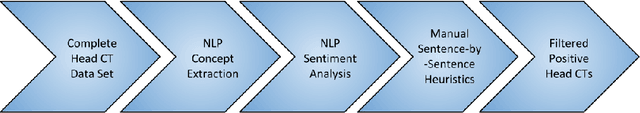
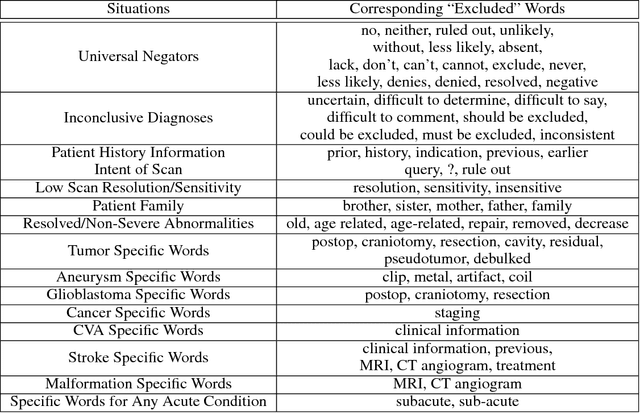
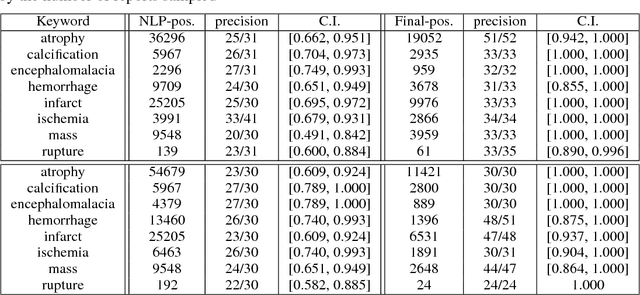
Abstract:Obtaining datasets labeled to facilitate model development is a challenge for most machine learning tasks. The difficulty is heightened for medical imaging, where data itself is limited in accessibility and labeling requires costly time and effort by trained medical specialists. Medical imaging studies, however, are often accompanied by a medical report produced by a radiologist, identifying important features on the corresponding scan for other physicians not specifically trained in radiology. We propose a methodology for approximating image-level labels for radiology studies from associated reports using a general purpose language processing tool for medical concept extraction and sentiment analysis, and simple manually crafted heuristics for false positive reduction. Using this approach, we label more than 175,000 Head CT studies for the presence of 33 features indicative of 11 clinically relevant conditions. For 27 of the 30 keywords that yielded positive results (3 had no occurrences), the lower bound of the confidence intervals created to estimate the percentage of accurately labeled reports was above 85%, with the average being above 95%. Though noisier then manual labeling, these results suggest this method to be a viable means of labeling medical images at scale.
Weakly Supervised Medical Diagnosis and Localization from Multiple Resolutions
Mar 21, 2018
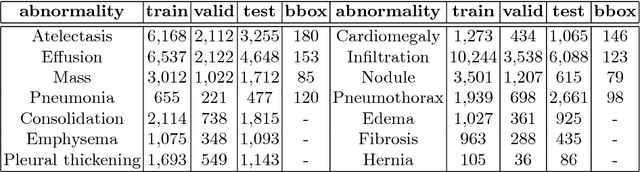
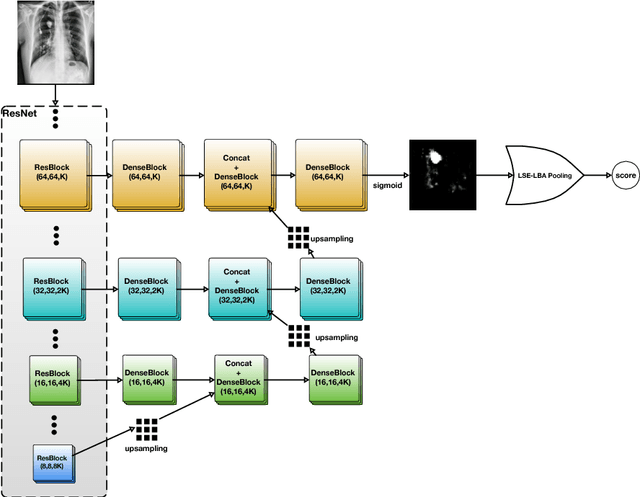
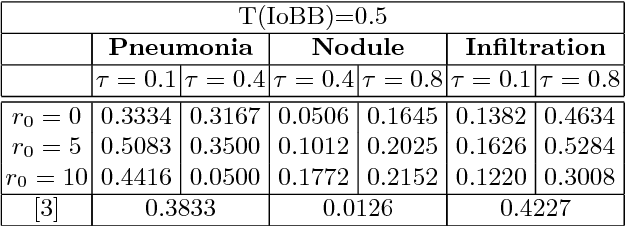
Abstract:Diagnostic imaging often requires the simultaneous identification of a multitude of findings of varied size and appearance. Beyond global indication of said findings, the prediction and display of localization information improves trust in and understanding of results when augmenting clinical workflow. Medical training data rarely includes more than global image-level labels as segmentations are time-consuming and expensive to collect. We introduce an approach to managing these practical constraints by applying a novel architecture which learns at multiple resolutions while generating saliency maps with weak supervision. Further, we parameterize the Log-Sum-Exp pooling function with a learnable lower-bounded adaptation (LSE-LBA) to build in a sharpness prior and better handle localizing abnormalities of different sizes using only image-level labels. Applying this approach to interpreting chest x-rays, we set the state of the art on 9 abnormalities in the NIH's CXR14 dataset while generating saliency maps with the highest resolution to date.
Learning to diagnose from scratch by exploiting dependencies among labels
Feb 01, 2018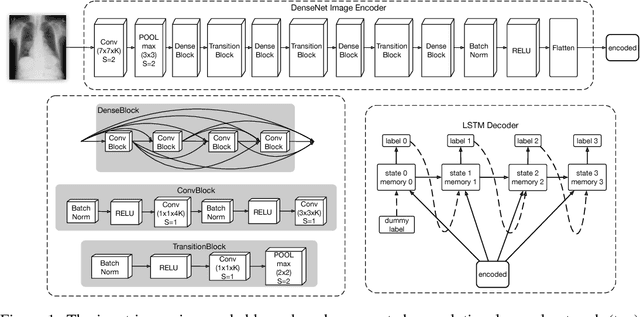

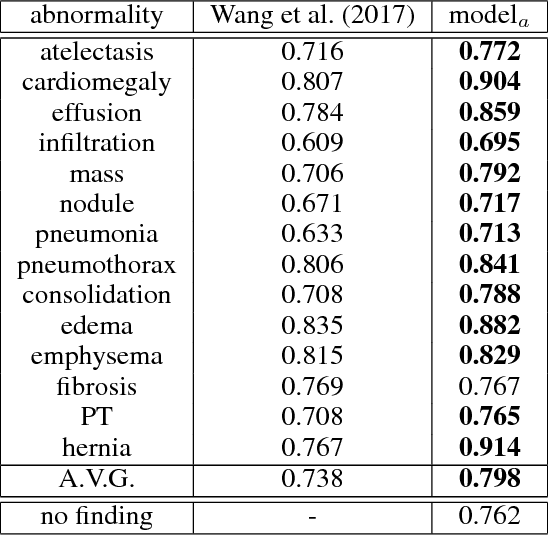
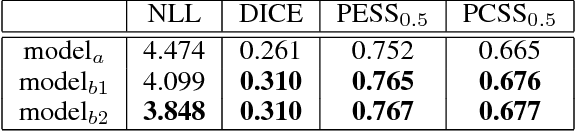
Abstract:The field of medical diagnostics contains a wealth of challenges which closely resemble classical machine learning problems; practical constraints, however, complicate the translation of these endpoints naively into classical architectures. Many tasks in radiology, for example, are largely problems of multi-label classification wherein medical images are interpreted to indicate multiple present or suspected pathologies. Clinical settings drive the necessity for high accuracy simultaneously across a multitude of pathological outcomes and greatly limit the utility of tools which consider only a subset. This issue is exacerbated by a general scarcity of training data and maximizes the need to extract clinically relevant features from available samples -- ideally without the use of pre-trained models which may carry forward undesirable biases from tangentially related tasks. We present and evaluate a partial solution to these constraints in using LSTMs to leverage interdependencies among target labels in predicting 14 pathologic patterns from chest x-rays and establish state of the art results on the largest publicly available chest x-ray dataset from the NIH without pre-training. Furthermore, we propose and discuss alternative evaluation metrics and their relevance in clinical practice.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge