Banban Huang
CoNIC Challenge: Pushing the Frontiers of Nuclear Detection, Segmentation, Classification and Counting
Mar 14, 2023



Abstract:Nuclear detection, segmentation and morphometric profiling are essential in helping us further understand the relationship between histology and patient outcome. To drive innovation in this area, we setup a community-wide challenge using the largest available dataset of its kind to assess nuclear segmentation and cellular composition. Our challenge, named CoNIC, stimulated the development of reproducible algorithms for cellular recognition with real-time result inspection on public leaderboards. We conducted an extensive post-challenge analysis based on the top-performing models using 1,658 whole-slide images of colon tissue. With around 700 million detected nuclei per model, associated features were used for dysplasia grading and survival analysis, where we demonstrated that the challenge's improvement over the previous state-of-the-art led to significant boosts in downstream performance. Our findings also suggest that eosinophils and neutrophils play an important role in the tumour microevironment. We release challenge models and WSI-level results to foster the development of further methods for biomarker discovery.
ConvNeXt-backbone HoVerNet for nuclei segmentation and classification
Mar 29, 2022
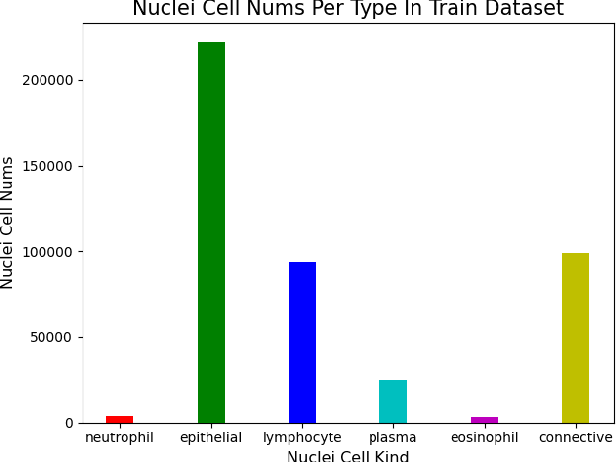


Abstract:This manuscript gives a brief description of the algorithm used to participate in CoNIC Challenge 2022. After the baseline was made available, we follow the method in it and replace the ResNet baseline with ConvNeXt one. Moreover, we propose to first convert RGB space to Haematoxylin-Eosin-DAB(HED) space, then use Haematoxylin composition of origin image to smooth semantic one hot label. Afterwards, nuclei distribution of train and valid set are explored to select the best fold split for training model for final test phase submission. Results on validation set shows that even with channel of each stage smaller in number, HoVerNet with ConvNeXt-tiny backbone still improves the mPQ+ by 0.04 and multi r2 by 0.0144
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge