Ayisha Imran
Uni-Hema: Unified Model for Digital Hematopathology
Nov 19, 2025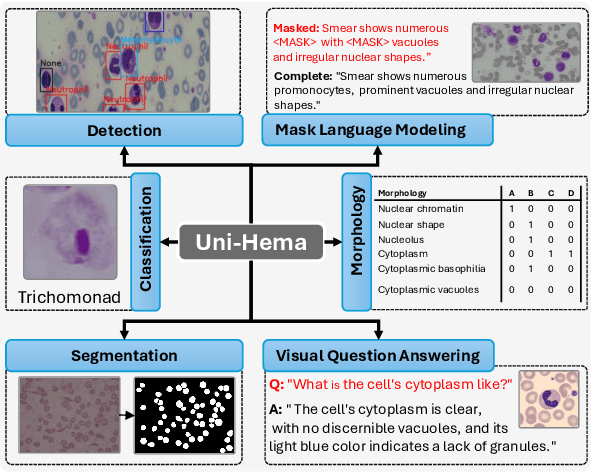
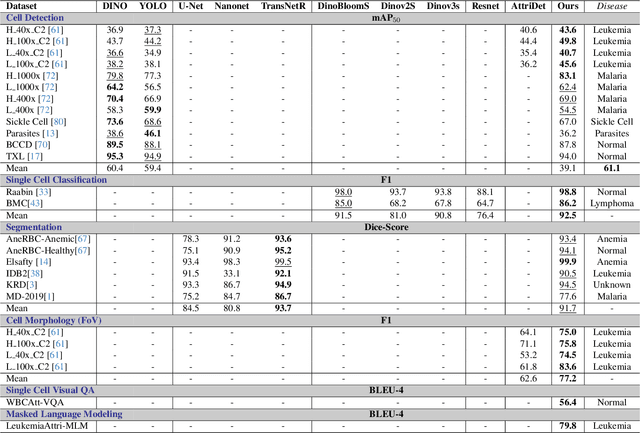
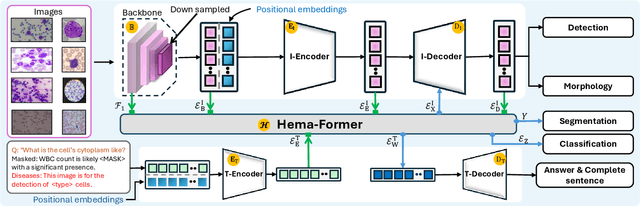
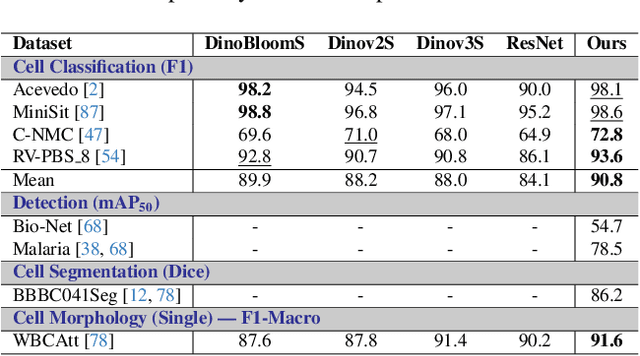
Abstract:Digital hematopathology requires cell-level analysis across diverse disease categories, including malignant disorders (e.g., leukemia), infectious conditions (e.g., malaria), and non-malignant red blood cell disorders (e.g., sickle cell disease). Whether single-task, vision-language, WSI-optimized, or single-cell hematology models, these approaches share a key limitation, they cannot provide unified, multi-task, multi-modal reasoning across the complexities of digital hematopathology. To overcome these limitations, we propose Uni-Hema, a multi-task, unified model for digital hematopathology integrating detection, classification, segmentation, morphology prediction, and reasoning across multiple diseases. Uni-Hema leverages 46 publicly available datasets, encompassing over 700K images and 21K question-answer pairs, and is built upon Hema-Former, a multimodal module that bridges visual and textual representations at the hierarchy level for the different tasks (detection, classification, segmentation, morphology, mask language modeling and visual question answer) at different granularity. Extensive experiments demonstrate that Uni-Hema achieves comparable or superior performance to train on a single-task and single dataset models, across diverse hematological tasks, while providing interpretable, morphologically relevant insights at the single-cell level. Our framework establishes a new standard for multi-task and multi-modal digital hematopathology. The code will be made publicly available.
Leveraging Sparse Annotations for Leukemia Diagnosis on the Large Leukemia Dataset
Apr 03, 2025Abstract:Leukemia is 10th most frequently diagnosed cancer and one of the leading causes of cancer related deaths worldwide. Realistic analysis of Leukemia requires White Blook Cells (WBC) localization, classification, and morphological assessment. Despite deep learning advances in medical imaging, leukemia analysis lacks a large, diverse multi-task dataset, while existing small datasets lack domain diversity, limiting real world applicability. To overcome dataset challenges, we present a large scale WBC dataset named Large Leukemia Dataset (LLD) and novel methods for detecting WBC with their attributes. Our contribution here is threefold. First, we present a large-scale Leukemia dataset collected through Peripheral Blood Films (PBF) from several patients, through multiple microscopes, multi cameras, and multi magnification. To enhance diagnosis explainability and medical expert acceptance, each leukemia cell is annotated at 100x with 7 morphological attributes, ranging from Cell Size to Nuclear Shape. Secondly, we propose a multi task model that not only detects WBCs but also predicts their attributes, providing an interpretable and clinically meaningful solution. Third, we propose a method for WBC detection with attribute analysis using sparse annotations. This approach reduces the annotation burden on hematologists, requiring them to mark only a small area within the field of view. Our method enables the model to leverage the entire field of view rather than just the annotated regions, enhancing learning efficiency and diagnostic accuracy. From diagnosis explainability to overcoming domain shift challenges, presented datasets could be used for many challenging aspects of microscopic image analysis. The datasets, code, and demo are available at: https://im.itu.edu.pk/sparse-leukemiaattri/
A Large-scale Multi Domain Leukemia Dataset for the White Blood Cells Detection with Morphological Attributes for Explainability
May 17, 2024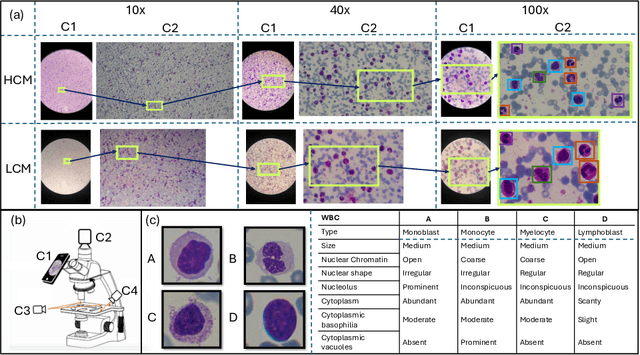
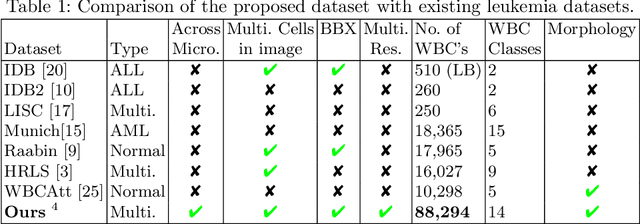
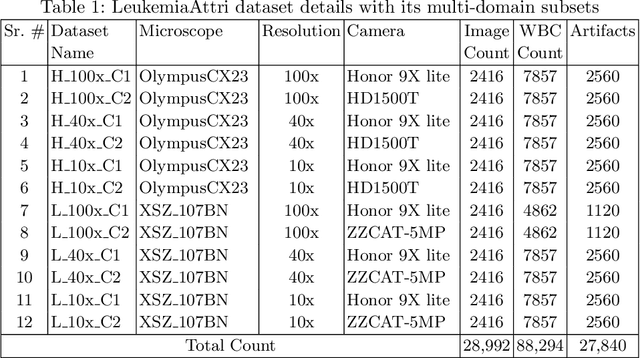
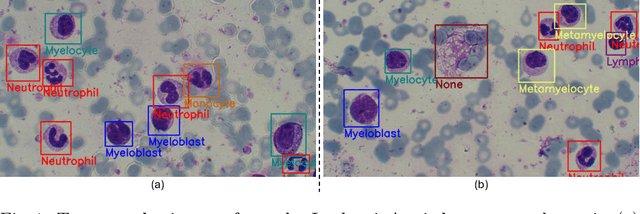
Abstract:Earlier diagnosis of Leukemia can save thousands of lives annually. The prognosis of leukemia is challenging without the morphological information of White Blood Cells (WBC) and relies on the accessibility of expensive microscopes and the availability of hematologists to analyze Peripheral Blood Samples (PBS). Deep Learning based methods can be employed to assist hematologists. However, these algorithms require a large amount of labeled data, which is not readily available. To overcome this limitation, we have acquired a realistic, generalized, and large dataset. To collect this comprehensive dataset for real-world applications, two microscopes from two different cost spectrums (high-cost HCM and low-cost LCM) are used for dataset capturing at three magnifications (100x, 40x, 10x) through different sensors (high-end camera for HCM, middle-level camera for LCM and mobile-phone camera for both). The high-sensor camera is 47 times more expensive than the middle-level camera and HCM is 17 times more expensive than LCM. In this collection, using HCM at high resolution (100x), experienced hematologists annotated 10.3k WBC types (14) and artifacts, having 55k morphological labels (Cell Size, Nuclear Chromatin, Nuclear Shape, etc.) from 2.4k images of several PBS leukemia patients. Later on, these annotations are transferred to other 2 magnifications of HCM, and 3 magnifications of LCM, and on each camera captured images. Along with the LeukemiaAttri dataset, we provide baselines over multiple object detectors and Unsupervised Domain Adaptation (UDA) strategies, along with morphological information-based attribute prediction. The dataset will be publicly available after publication to facilitate the research in this direction.
A Dataset and Benchmark for Malaria Life-Cycle Classification in Thin Blood Smear Images
Feb 17, 2021



Abstract:Malaria microscopy, microscopic examination of stained blood slides to detect parasite Plasmodium, is considered to be a gold-standard for detecting life-threatening disease malaria. Detecting the plasmodium parasite requires a skilled examiner and may take up to 10 to 15 minutes to completely go through the whole slide. Due to a lack of skilled medical professionals in the underdeveloped or resource deficient regions, many cases go misdiagnosed; resulting in unavoidable complications and/or undue medication. We propose to complement the medical professionals by creating a deep learning-based method to automatically detect (localize) the plasmodium parasites in the photograph of stained film. To handle the unbalanced nature of the dataset, we adopt a two-stage approach. Where the first stage is trained to detect blood cells and classify them into just healthy or infected. The second stage is trained to classify each detected cell further into the life-cycle stage. To facilitate the research in machine learning-based malaria microscopy, we introduce a new large scale microscopic image malaria dataset. Thirty-eight thousand cells are tagged from the 345 microscopic images of different Giemsa-stained slides of blood samples. Extensive experimentation is performed using different CNN backbones including VGG, DenseNet, and ResNet on this dataset. Our experiments and analysis reveal that the two-stage approach works better than the one-stage approach for malaria detection. To ensure the usability of our approach, we have also developed a mobile app that will be used by local hospitals for investigation and educational purposes. The dataset, its annotations, and implementation codes will be released upon publication of the paper.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge