Ashraf Mahgoub
Lerna: Transformer Architectures for Configuring Error Correction Tools for Short- and Long-Read Genome Sequencing
Dec 19, 2021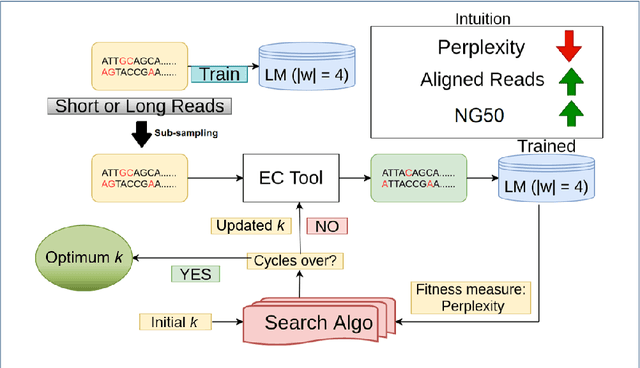
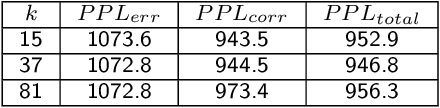
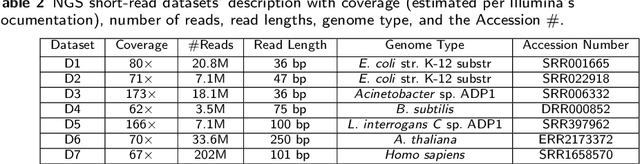
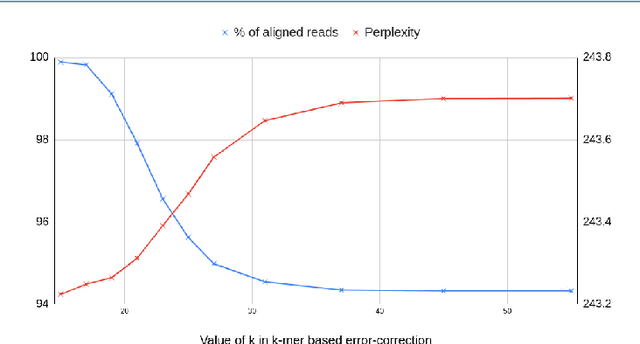
Abstract:Sequencing technologies are prone to errors, making error correction (EC) necessary for downstream applications. EC tools need to be manually configured for optimal performance. We find that the optimal parameters (e.g., k-mer size) are both tool- and dataset-dependent. Moreover, evaluating the performance (i.e., Alignment-rate or Gain) of a given tool usually relies on a reference genome, but quality reference genomes are not always available. We introduce Lerna for the automated configuration of k-mer-based EC tools. Lerna first creates a language model (LM) of the uncorrected genomic reads; then, calculates the perplexity metric to evaluate the corrected reads for different parameter choices. Next, it finds the one that produces the highest alignment rate without using a reference genome. The fundamental intuition of our approach is that the perplexity metric is inversely correlated with the quality of the assembly after error correction. Results: First, we show that the best k-mer value can vary for different datasets, even for the same EC tool. Second, we show the gains of our LM using its component attention-based transformers. We show the model's estimation of the perplexity metric before and after error correction. The lower the perplexity after correction, the better the k-mer size. We also show that the alignment rate and assembly quality computed for the corrected reads are strongly negatively correlated with the perplexity, enabling the automated selection of k-mer values for better error correction, and hence, improved assembly quality. Additionally, we show that our attention-based models have significant runtime improvement for the entire pipeline -- 18X faster than previous works, due to parallelizing the attention mechanism and the use of JIT compilation for GPU inferencing.
JANUS: Benchmarking Commercial and Open-Source Cloud and Edge Platforms for Object and Anomaly Detection Workloads
Dec 09, 2020



Abstract:With diverse IoT workloads, placing compute and analytics close to where data is collected is becoming increasingly important. We seek to understand what is the performance and the cost implication of running analytics on IoT data at the various available platforms. These workloads can be compute-light, such as outlier detection on sensor data, or compute-intensive, such as object detection from video feeds obtained from drones. In our paper, JANUS, we profile the performance/$ and the compute versus communication cost for a compute-light IoT workload and a compute-intensive IoT workload. In addition, we also look at the pros and cons of some of the proprietary deep-learning object detection packages, such as Amazon Rekognition, Google Vision, and Azure Cognitive Services, to contrast with open-source and tunable solutions, such as Faster R-CNN (FRCNN). We find that AWS IoT Greengrass delivers at least 2X lower latency and 1.25X lower cost compared to all other cloud platforms for the compute-light outlier detection workload. For the compute-intensive streaming video analytics task, an opensource solution to object detection running on cloud VMs saves on dollar costs compared to proprietary solutions provided by Amazon, Microsoft, and Google, but loses out on latency (up to 6X). If it runs on a low-powered edge device, the latency is up to 49X lower.
* Appeared at the IEEE Cloud 2020 conference. 10 pages
ATHENA: Automated Tuning of Genomic Error Correction Algorithms using Language Models
Dec 30, 2018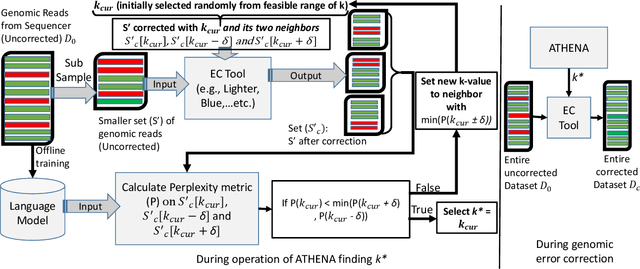

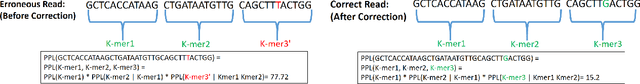
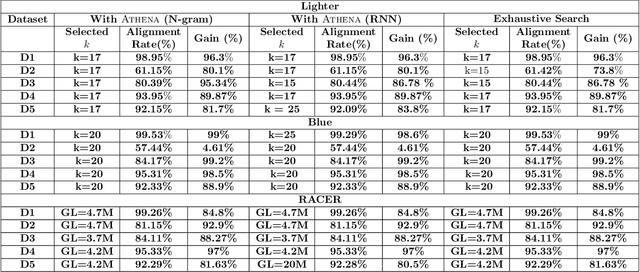
Abstract:The performance of most error-correction algorithms that operate on genomic sequencer reads is dependent on the proper choice of its configuration parameters, such as the value of k in k-mer based techniques. In this work, we target the problem of finding the best values of these configuration parameters to optimize error correction. We perform this in a data-driven manner, due to the observation that different configuration parameters are optimal for different datasets, i.e., from different instruments and organisms. We use language modeling techniques from the Natural Language Processing (NLP) domain in our algorithmic suite, Athena, to automatically tune the performance-sensitive configuration parameters. Through the use of N-Gram and Recurrent Neural Network (RNN) language modeling, we validate the intuition that the EC performance can be computed quantitatively and efficiently using the perplexity metric, prevalent in NLP. After training the language model, we show that the perplexity metric calculated for runtime data has a strong negative correlation with the correction of the erroneous NGS reads. Therefore, we use the perplexity metric to guide a hill climbing-based search, converging toward the best $k$-value. Our approach is suitable for both de novo and comparative sequencing (resequencing), eliminating the need for a reference genome to serve as the ground truth. This is important because the use of a reference genome often carries forward the biases along the stages of the pipeline.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge