Ananya Jana
Comparative Analysis of OpenAI GPT-4o and DeepSeek R1 for Scientific Text Categorization Using Prompt Engineering
Mar 03, 2025



Abstract:This study examines how large language models categorize sentences from scientific papers using prompt engineering. We use two advanced web-based models, GPT-4o (by OpenAI) and DeepSeek R1, to classify sentences into predefined relationship categories. DeepSeek R1 has been tested on benchmark datasets in its technical report. However, its performance in scientific text categorization remains unexplored. To address this gap, we introduce a new evaluation method designed specifically for this task. We also compile a dataset of cleaned scientific papers from diverse domains. This dataset provides a platform for comparing the two models. Using this dataset, we analyze their effectiveness and consistency in categorization.
Evaluating the Suitability of Different Intraoral Scan Resolutions for Deep Learning-Based Tooth Segmentation
Feb 26, 2025Abstract:Intraoral scans are widely used in digital dentistry for tasks such as dental restoration, treatment planning, and orthodontic procedures. These scans contain detailed topological information, but manual annotation of these scans remains a time-consuming task. Deep learning-based methods have been developed to automate tasks such as tooth segmentation. A typical intraoral scan contains over 200,000 mesh cells, making direct processing computationally expensive. Models are often trained on downsampled versions, typically with 10,000 or 16,000 cells. Previous studies suggest that downsampling may degrade segmentation accuracy, but the extent of this degradation remains unclear. Understanding the extent of degradation is crucial for deploying ML models on edge devices. This study evaluates the extent of performance degradation with decreasing resolution. We train a deep learning model (PointMLP) on intraoral scans decimated to 16K, 10K, 8K, 6K, 4K, and 2K mesh cells. Models trained at lower resolutions are tested on high-resolution scans to assess performance. Our goal is to identify a resolution that balances computational efficiency and segmentation accuracy.
A Critical Analysis of the Limitation of Deep Learning based 3D Dental Mesh Segmentation Methods in Segmenting Partial Scans
Apr 29, 2023Abstract:Tooth segmentation from intraoral scans is a crucial part of digital dentistry. Many Deep Learning based tooth segmentation algorithms have been developed for this task. In most of the cases, high accuracy has been achieved, although, most of the available tooth segmentation techniques make an implicit restrictive assumption of full jaw model and they report accuracy based on full jaw models. Medically, however, in certain cases, full jaw tooth scan is not required or may not be available. Given this practical issue, it is important to understand the robustness of currently available widely used Deep Learning based tooth segmentation techniques. For this purpose, we applied available segmentation techniques on partial intraoral scans and we discovered that the available deep Learning techniques under-perform drastically. The analysis and comparison presented in this work would help us in understanding the severity of the problem and allow us to develop robust tooth segmentation technique without strong assumption of full jaw model.
3D Tooth Mesh Segmentation with Simplified Mesh Cell Representation
Jan 25, 2023Abstract:Manual tooth segmentation of 3D tooth meshes is tedious and there is variations among dentists. %Manual tooth annotation of 3D tooth meshes is a tedious task. Several deep learning based methods have been proposed to perform automatic tooth mesh segmentation. Many of the proposed tooth mesh segmentation algorithms summarize the mesh cell as - the cell center or barycenter, the normal at barycenter, the cell vertices and the normals at the cell vertices. Summarizing of the mesh cell/triangle in this manner imposes an implicit structural constraint and makes it difficult to work with multiple resolutions which is done in many point cloud based deep learning algorithms. We propose a novel segmentation method which utilizes only the barycenter and the normal at the barycenter information of the mesh cell and yet achieves competitive performance. We are the first to demonstrate that it is possible to relax the implicit structural constraint and yet achieve superior segmentation performance
Automatic Tooth Segmentation from 3D Dental Model using Deep Learning: A Quantitative Analysis of what can be learnt from a Single 3D Dental Model
Sep 16, 2022



Abstract:3D tooth segmentation is an important task for digital orthodontics. Several Deep Learning methods have been proposed for automatic tooth segmentation from 3D dental models or intraoral scans. These methods require annotated 3D intraoral scans. Manually annotating 3D intraoral scans is a laborious task. One approach is to devise self-supervision methods to reduce the manual labeling effort. Compared to other types of point cloud data like scene point cloud or shape point cloud data, 3D tooth point cloud data has a very regular structure and a strong shape prior. We look at how much representative information can be learnt from a single 3D intraoral scan. We evaluate this quantitatively with the help of ten different methods of which six are generic point cloud segmentation methods whereas the other four are tooth segmentation specific methods. Surprisingly, we find that with a single 3D intraoral scan training, the Dice score can be as high as 0.86 whereas the full training set gives Dice score of 0.94. We conclude that the segmentation methods can learn a great deal of information from a single 3D tooth point cloud scan under suitable conditions e.g. data augmentation. We are the first to quantitatively evaluate and demonstrate the representation learning capability of Deep Learning methods from a single 3D intraoral scan. This can enable building self-supervision methods for tooth segmentation under extreme data limitation scenario by leveraging the available data to the fullest possible extent.
Global and Local Interpretation of black-box Machine Learning models to determine prognostic factors from early COVID-19 data
Sep 10, 2021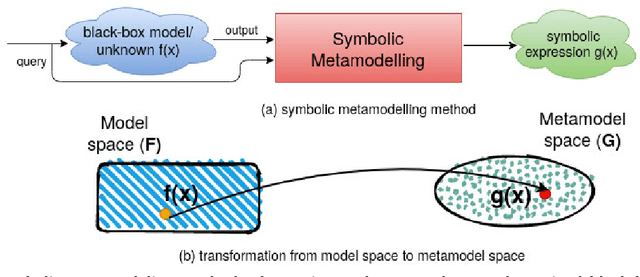



Abstract:The COVID-19 corona virus has claimed 4.1 million lives, as of July 24, 2021. A variety of machine learning models have been applied to related data to predict important factors such as the severity of the disease, infection rate and discover important prognostic factors. Often the usefulness of the findings from the use of these techniques is reduced due to lack of method interpretability. Some recent progress made on the interpretability of machine learning models has the potential to unravel more insights while using conventional machine learning models. In this work, we analyze COVID-19 blood work data with some of the popular machine learning models; then we employ state-of-the-art post-hoc local interpretability techniques(e.g.- SHAP, LIME), and global interpretability techniques(e.g. - symbolic metamodeling) to the trained black-box models to draw interpretable conclusions. In the gamut of machine learning algorithms, regressions remain one of the simplest and most explainable models with clear mathematical formulation. We explore one of the most recent techniques called symbolic metamodeling to find the mathematical expression of the machine learning models for COVID-19. We identify Acute Kidney Injury (AKI), initial Albumin level (ALBI), Aspartate aminotransferase (ASTI), Total Bilirubin initial(TBILI) and D-Dimer initial (DIMER) as major prognostic factors of the disease severity. Our contributions are- (i) uncover the underlying mathematical expression for the black-box models on COVID-19 severity prediction task (ii) we are the first to apply symbolic metamodeling to this task, and (iii) discover important features and feature interactions.
Liver Fibrosis and NAS scoring from CT images using self-supervised learning and texture encoding
Mar 15, 2021



Abstract:Non-alcoholic fatty liver disease (NAFLD) is one of the most common causes of chronic liver diseases (CLD) which can progress to liver cancer. The severity and treatment of NAFLD is determined by NAFLD Activity Scores (NAS)and liver fibrosis stage, which are usually obtained from liver biopsy. However, biopsy is invasive in nature and involves risk of procedural complications. Current methods to predict the fibrosis and NAS scores from noninvasive CT images rely heavily on either a large annotated dataset or transfer learning using pretrained networks. However, the availability of a large annotated dataset cannot be always ensured andthere can be domain shifts when using transfer learning. In this work, we propose a self-supervised learning method to address both problems. As the NAFLD causes changes in the liver texture, we also propose to use texture encoded inputs to improve the performance of the model. Given a relatively small dataset with 30 patients, we employ a self-supervised network which achieves better performance than a network trained via transfer learning. The code is publicly available at https://github.com/ananyajana/fibrosis_code.
Deep Learning based NAS Score and Fibrosis Stage Prediction from CT and Pathology Data
Sep 22, 2020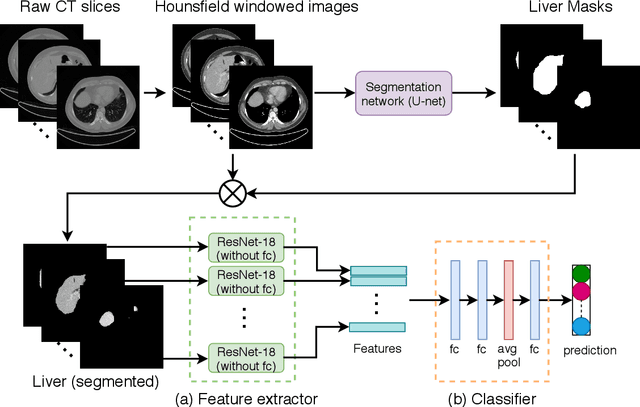
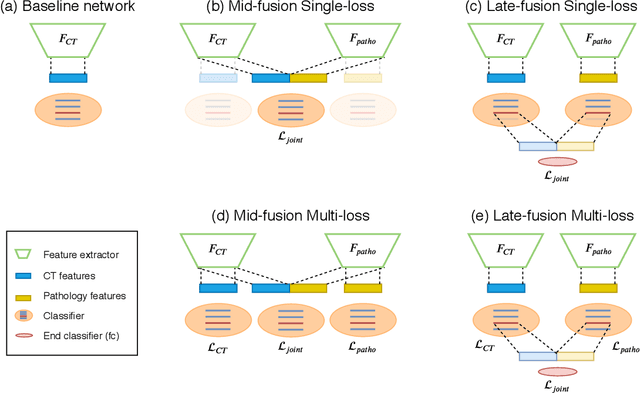


Abstract:Non-Alcoholic Fatty Liver Disease (NAFLD) is becoming increasingly prevalent in the world population. Without diagnosis at the right time, NAFLD can lead to non-alcoholic steatohepatitis (NASH) and subsequent liver damage. The diagnosis and treatment of NAFLD depend on the NAFLD activity score (NAS) and the liver fibrosis stage, which are usually evaluated from liver biopsies by pathologists. In this work, we propose a novel method to automatically predict NAS score and fibrosis stage from CT data that is non-invasive and inexpensive to obtain compared with liver biopsy. We also present a method to combine the information from CT and H\&E stained pathology data to improve the performance of NAS score and fibrosis stage prediction, when both types of data are available. This is of great value to assist the pathologists in computer-aided diagnosis process. Experiments on a 30-patient dataset illustrate the effectiveness of our method.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge