Vinod Rustgi
Global and Local Interpretation of black-box Machine Learning models to determine prognostic factors from early COVID-19 data
Sep 10, 2021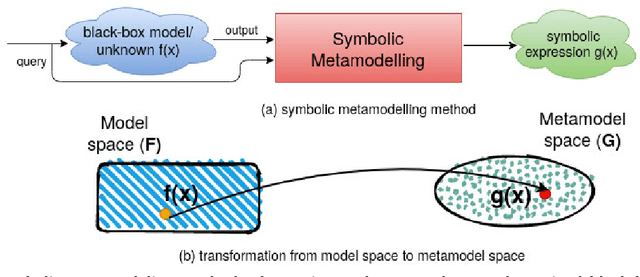



Abstract:The COVID-19 corona virus has claimed 4.1 million lives, as of July 24, 2021. A variety of machine learning models have been applied to related data to predict important factors such as the severity of the disease, infection rate and discover important prognostic factors. Often the usefulness of the findings from the use of these techniques is reduced due to lack of method interpretability. Some recent progress made on the interpretability of machine learning models has the potential to unravel more insights while using conventional machine learning models. In this work, we analyze COVID-19 blood work data with some of the popular machine learning models; then we employ state-of-the-art post-hoc local interpretability techniques(e.g.- SHAP, LIME), and global interpretability techniques(e.g. - symbolic metamodeling) to the trained black-box models to draw interpretable conclusions. In the gamut of machine learning algorithms, regressions remain one of the simplest and most explainable models with clear mathematical formulation. We explore one of the most recent techniques called symbolic metamodeling to find the mathematical expression of the machine learning models for COVID-19. We identify Acute Kidney Injury (AKI), initial Albumin level (ALBI), Aspartate aminotransferase (ASTI), Total Bilirubin initial(TBILI) and D-Dimer initial (DIMER) as major prognostic factors of the disease severity. Our contributions are- (i) uncover the underlying mathematical expression for the black-box models on COVID-19 severity prediction task (ii) we are the first to apply symbolic metamodeling to this task, and (iii) discover important features and feature interactions.
Liver Fibrosis and NAS scoring from CT images using self-supervised learning and texture encoding
Mar 15, 2021



Abstract:Non-alcoholic fatty liver disease (NAFLD) is one of the most common causes of chronic liver diseases (CLD) which can progress to liver cancer. The severity and treatment of NAFLD is determined by NAFLD Activity Scores (NAS)and liver fibrosis stage, which are usually obtained from liver biopsy. However, biopsy is invasive in nature and involves risk of procedural complications. Current methods to predict the fibrosis and NAS scores from noninvasive CT images rely heavily on either a large annotated dataset or transfer learning using pretrained networks. However, the availability of a large annotated dataset cannot be always ensured andthere can be domain shifts when using transfer learning. In this work, we propose a self-supervised learning method to address both problems. As the NAFLD causes changes in the liver texture, we also propose to use texture encoded inputs to improve the performance of the model. Given a relatively small dataset with 30 patients, we employ a self-supervised network which achieves better performance than a network trained via transfer learning. The code is publicly available at https://github.com/ananyajana/fibrosis_code.
Deep Learning based NAS Score and Fibrosis Stage Prediction from CT and Pathology Data
Sep 22, 2020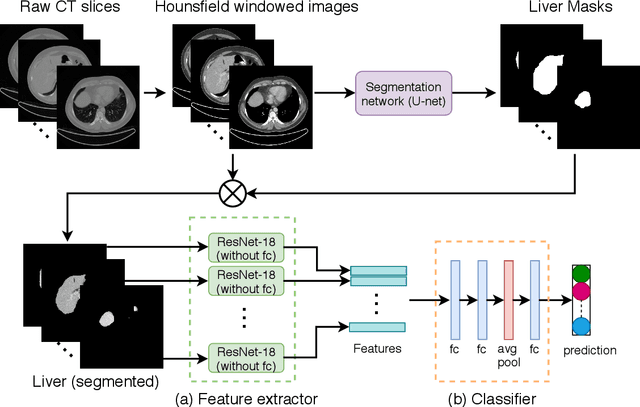
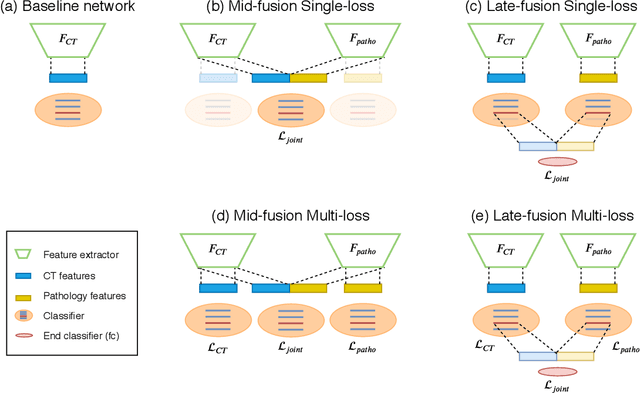


Abstract:Non-Alcoholic Fatty Liver Disease (NAFLD) is becoming increasingly prevalent in the world population. Without diagnosis at the right time, NAFLD can lead to non-alcoholic steatohepatitis (NASH) and subsequent liver damage. The diagnosis and treatment of NAFLD depend on the NAFLD activity score (NAS) and the liver fibrosis stage, which are usually evaluated from liver biopsies by pathologists. In this work, we propose a novel method to automatically predict NAS score and fibrosis stage from CT data that is non-invasive and inexpensive to obtain compared with liver biopsy. We also present a method to combine the information from CT and H\&E stained pathology data to improve the performance of NAS score and fibrosis stage prediction, when both types of data are available. This is of great value to assist the pathologists in computer-aided diagnosis process. Experiments on a 30-patient dataset illustrate the effectiveness of our method.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge