Amir Ahooye Atashin
Variational Leakage: The Role of Information Complexity in Privacy Leakage
Jun 08, 2021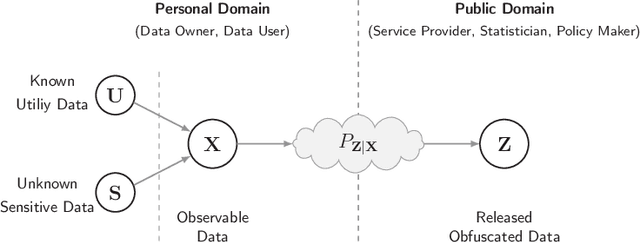
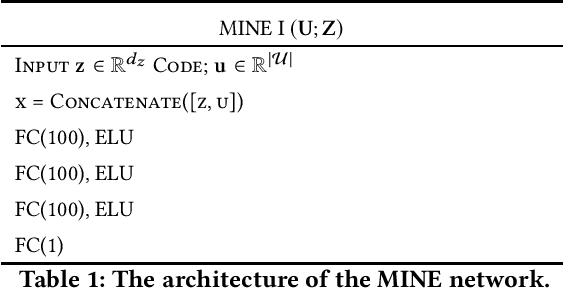
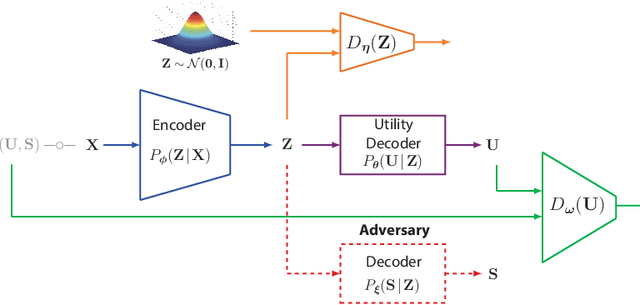
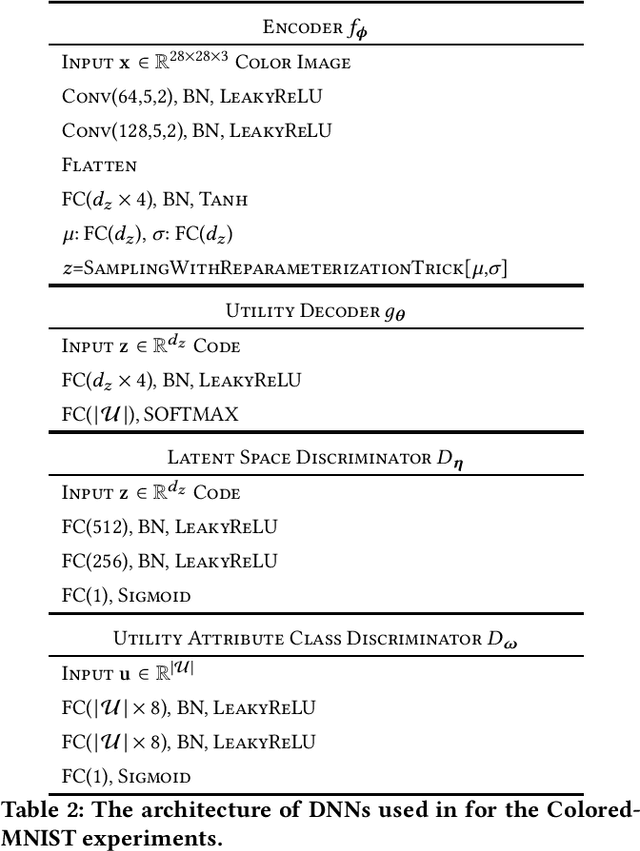
Abstract:We study the role of information complexity in privacy leakage about an attribute of an adversary's interest, which is not known a priori to the system designer. Considering the supervised representation learning setup and using neural networks to parameterize the variational bounds of information quantities, we study the impact of the following factors on the amount of information leakage: information complexity regularizer weight, latent space dimension, the cardinalities of the known utility and unknown sensitive attribute sets, the correlation between utility and sensitive attributes, and a potential bias in a sensitive attribute of adversary's interest. We conduct extensive experiments on Colored-MNIST and CelebA datasets to evaluate the effect of information complexity on the amount of intrinsic leakage.
An efficient projection neural network for $\ell_1$-regularized logistic regression
May 12, 2021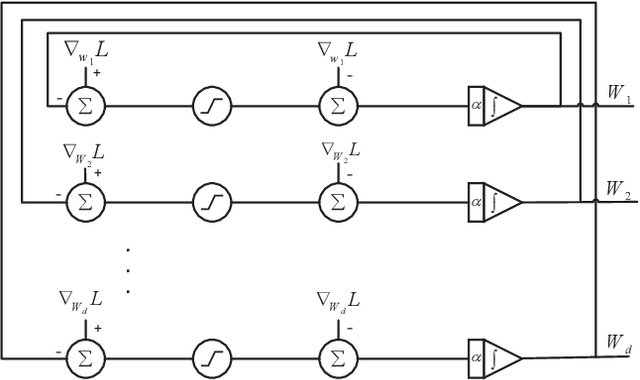
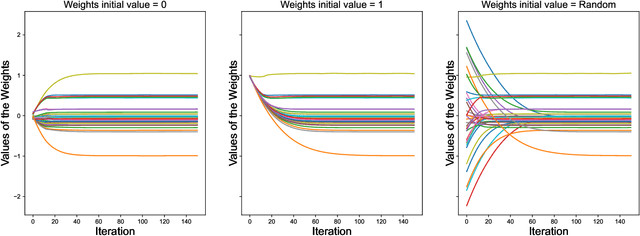
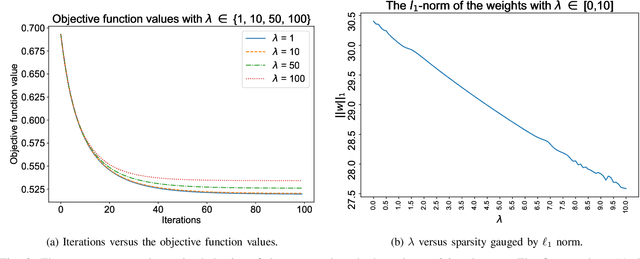
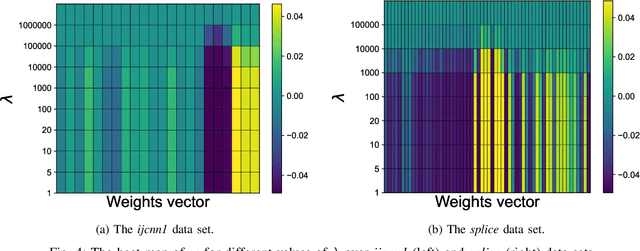
Abstract:$\ell_1$ regularization has been used for logistic regression to circumvent the overfitting and use the estimated sparse coefficient for feature selection. However, the challenge of such a regularization is that the $\ell_1$ norm is not differentiable, making the standard algorithms for convex optimization not applicable to this problem. This paper presents a simple projection neural network for $\ell_1$-regularized logistics regression. In contrast to many available solvers in the literature, the proposed neural network does not require any extra auxiliary variable nor any smooth approximation, and its complexity is almost identical to that of the gradient descent for logistic regression without $\ell_1$ regularization, thanks to the projection operator. We also investigate the convergence of the proposed neural network by using the Lyapunov theory and show that it converges to a solution of the problem with any arbitrary initial value. The proposed neural solution significantly outperforms state-of-the-art methods with respect to the execution time and is competitive in terms of accuracy and AUROC.
SANOM Results for OAEI 2019
Jun 09, 2020
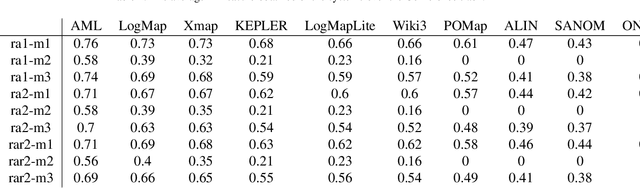

Abstract:Simulated annealing-based ontology matching (SANOM) participates for the second time at the ontology alignment evaluation initiative (OAEI) 2019. This paper contains the configuration of SANOM and its results on the anatomy and conference tracks. In comparison to the OAEI 2017, SANOM has improved significantly, and its results are competitive with the state-of-the-art systems. In particular, SANOM has the highest recall rate among the participated systems in the conference track, and is competitive with AML, the best performing system, in terms of F-measure. SANOM is also competitive with LogMap on the anatomy track, which is the best performing system in this track with no usage of particular biomedical background knowledge. SANOM has been adapted to the HOBBIT platfrom and is now available for the registered users.
Comparison of ontology alignment systems across single matching task via the McNemar's test
Apr 20, 2018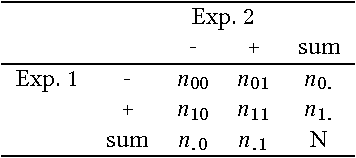
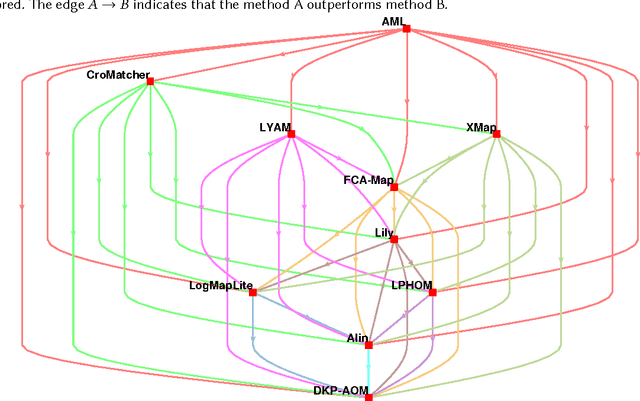
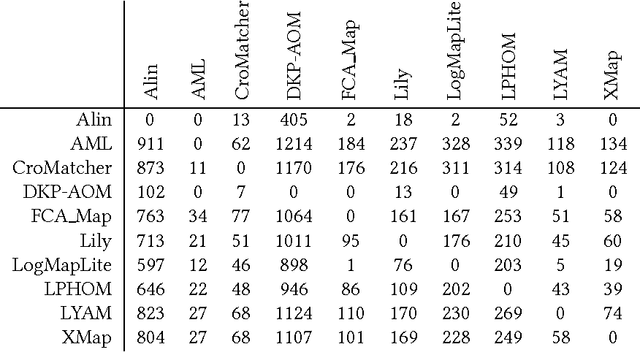
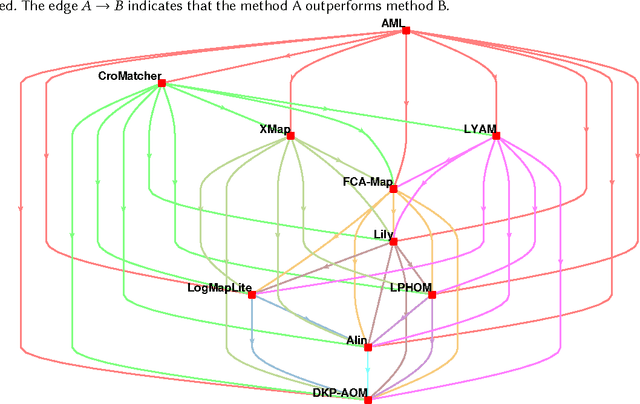
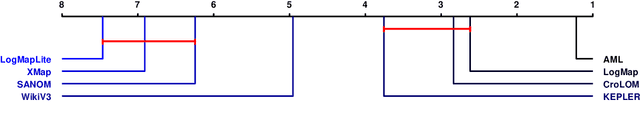
Abstract:Ontology alignment is widely-used to find the correspondences between different ontologies in diverse fields.After discovering the alignments,several performance scores are available to evaluate them.The scores typically require the identified alignment and a reference containing the underlying actual correspondences of the given ontologies.The current trend in the alignment evaluation is to put forward a new score(e.g., precision, weighted precision, etc.)and to compare various alignments by juxtaposing the obtained scores. However,it is substantially provocative to select one measure among others for comparison.On top of that, claiming if one system has a better performance than one another cannot be substantiated solely by comparing two scalars.In this paper,we propose the statistical procedures which enable us to theoretically favor one system over one another.The McNemar's test is the statistical means by which the comparison of two ontology alignment systems over one matching task is drawn.The test applies to a 2x2 contingency table which can be constructed in two different ways based on the alignments,each of which has their own merits/pitfalls.The ways of the contingency table construction and various apposite statistics from the McNemar's test are elaborated in minute detail.In the case of having more than two alignment systems for comparison, the family-wise error rate is expected to happen. Thus, the ways of preventing such an error are also discussed.A directed graph visualizes the outcome of the McNemar's test in the presence of multiple alignment systems.From this graph, it is readily understood if one system is better than one another or if their differences are imperceptible.The proposed statistical methodologies are applied to the systems participated in the OAEI 2016 anatomy track, and also compares several well-known similarity metrics for the same matching problem.
Training LDCRF model on unsegmented sequences using Connectionist Temporal Classification
Sep 06, 2016



Abstract:Many machine learning problems such as speech recognition, gesture recognition, and handwriting recognition are concerned with simultaneous segmentation and labeling of sequence data. Latent-dynamic conditional random field (LDCRF) is a well-known discriminative method that has been successfully used for this task. However, LDCRF can only be trained with pre-segmented data sequences in which the label of each frame is available apriori. In the realm of neural networks, the invention of connectionist temporal classification (CTC) made it possible to train recurrent neural networks on unsegmented sequences with great success. In this paper, we use CTC to train an LDCRF model on unsegmented sequences. Experimental results on two gesture recognition tasks show that the proposed method outperforms LDCRFs, hidden Markov models, and conditional random fields.
A two-stage learning method for protein-protein interaction prediction
Jul 18, 2016



Abstract:In this paper, a new method for PPI (proteinprotein interaction) prediction is proposed. In PPI prediction, a reliable and sufficient number of training samples is not available, but a large number of unlabeled samples is in hand. In the proposed method, the denoising auto encoders are employed for learning robust features. The obtained robust features are used in order to train a classifier with a better performance. The experimental results demonstrate the capabilities of the proposed method. Protein-protein interaction; Denoising auto encoder;Robust features; Unlabelled data;
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge