Alejandro Pazos
Application of Tabular Transformer Architectures for Operating System Fingerprinting
Feb 13, 2025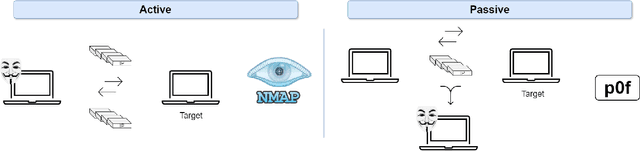

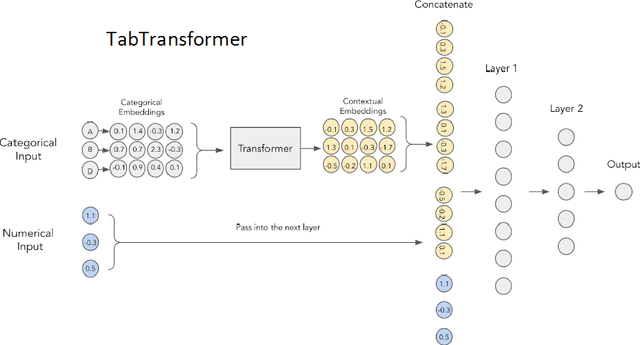
Abstract:Operating System (OS) fingerprinting is essential for network management and cybersecurity, enabling accurate device identification based on network traffic analysis. Traditional rule-based tools such as Nmap and p0f face challenges in dynamic environments due to frequent OS updates and obfuscation techniques. While Machine Learning (ML) approaches have been explored, Deep Learning (DL) models, particularly Transformer architectures, remain unexploited in this domain. This study investigates the application of Tabular Transformer architectures-specifically TabTransformer and FT-Transformer-for OS fingerprinting, leveraging structured network data from three publicly available datasets. Our experiments demonstrate that FT-Transformer generally outperforms traditional ML models, previous approaches and TabTransformer across multiple classification levels (OS family, major, and minor versions). The results establish a strong foundation for DL-based OS fingerprinting, improving accuracy and adaptability in complex network environments. Furthermore, we ensure the reproducibility of our research by providing an open-source implementation.
Machine Learning in management of precautionary closures caused by lipophilic biotoxins
Feb 14, 2024Abstract:Mussel farming is one of the most important aquaculture industries. The main risk to mussel farming is harmful algal blooms (HABs), which pose a risk to human consumption. In Galicia, the Spanish main producer of cultivated mussels, the opening and closing of the production areas is controlled by a monitoring program. In addition to the closures resulting from the presence of toxicity exceeding the legal threshold, in the absence of a confirmatory sampling and the existence of risk factors, precautionary closures may be applied. These decisions are made by experts without the support or formalisation of the experience on which they are based. Therefore, this work proposes a predictive model capable of supporting the application of precautionary closures. Achieving sensitivity, accuracy and kappa index values of 97.34%, 91.83% and 0.75 respectively, the kNN algorithm has provided the best results. This allows the creation of a system capable of helping in complex situations where forecast errors are more common.
Ensemble of Convolution Neural Networks on Heterogeneous Signals for Sleep Stage Scoring
Jul 23, 2021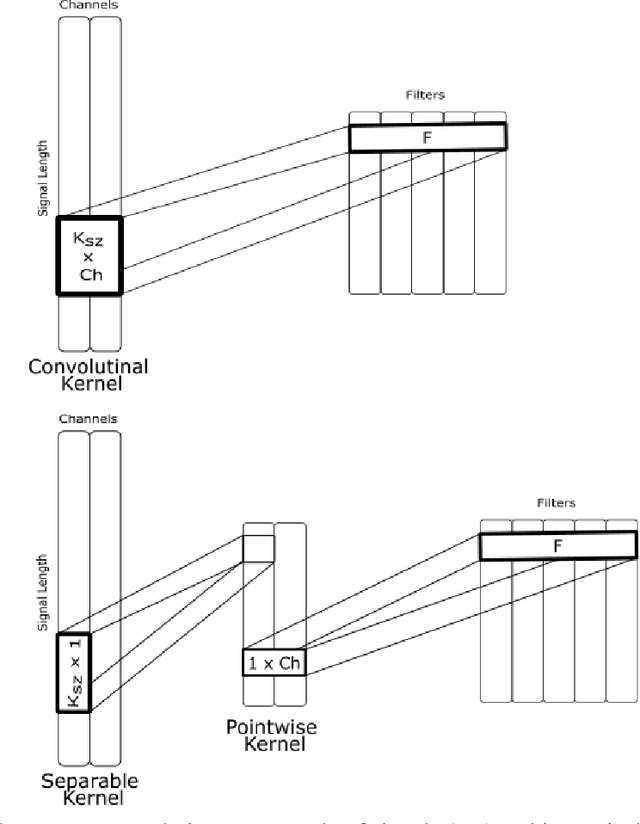

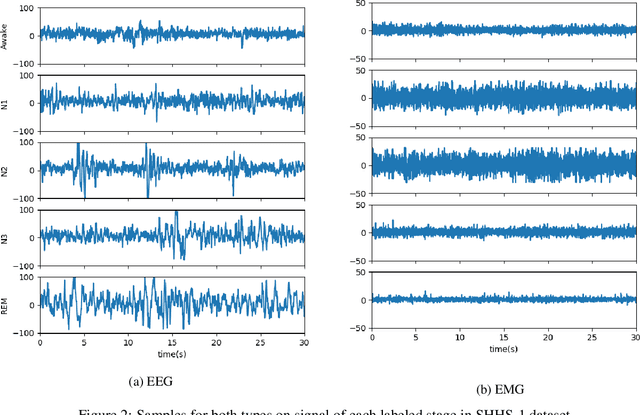
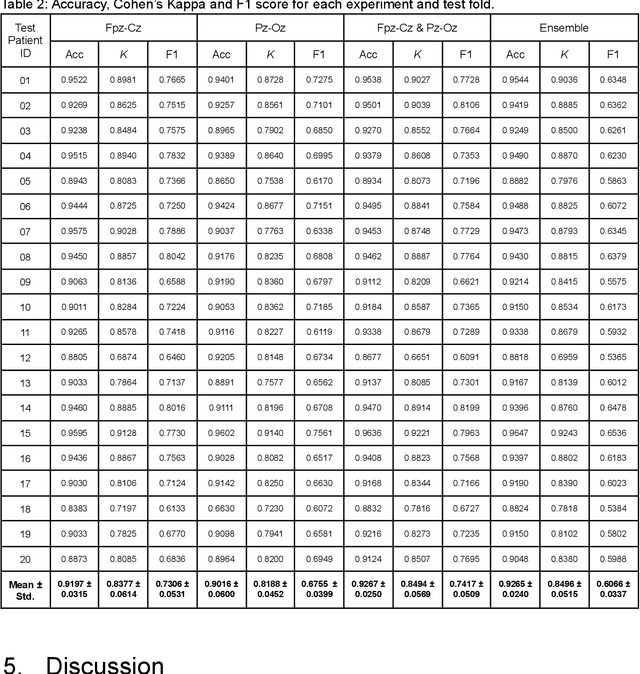
Abstract:Over the years, several approaches have tried to tackle the problem of performing an automatic scoring of the sleeping stages. Although any polysomnography usually collects over a dozen of different signals, this particular problem has been mainly tackled by using only the Electroencephalograms presented in those records. On the other hand, the other recorded signals have been mainly ignored by most works. This paper explores and compares the convenience of using additional signals apart from electroencephalograms. More specifically, this work uses the SHHS-1 dataset with 5,804 patients containing an electromyogram recorded simultaneously as two electroencephalograms. To compare the results, first, the same architecture has been evaluated with different input signals and all their possible combinations. These tests show how, using more than one signal especially if they are from different sources, improves the results of the classification. Additionally, the best models obtained for each combination of one or more signals have been used in ensemble models and, its performance has been compared showing the convenience of using these multi-signal models to improve the classification. The best overall model, an ensemble of Depth-wise Separational Convolutional Neural Networks, has achieved an accuracy of 86.06\% with a Cohen's Kappa of 0.80 and a $F_{1}$ of 0.77. Up to date, those are the best results on the complete dataset and it shows a significant improvement in the precision and recall for the most uncommon class in the dataset.
Automatic Assessment of Alzheimer's Disease Diagnosis Based on Deep Learning Techniques
May 18, 2021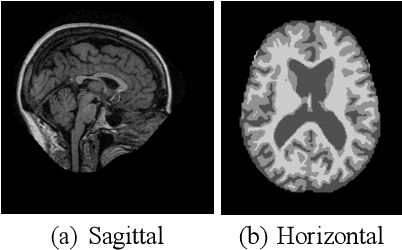
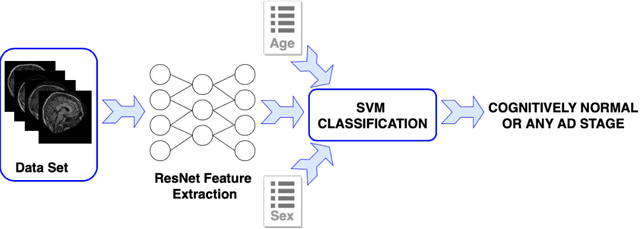
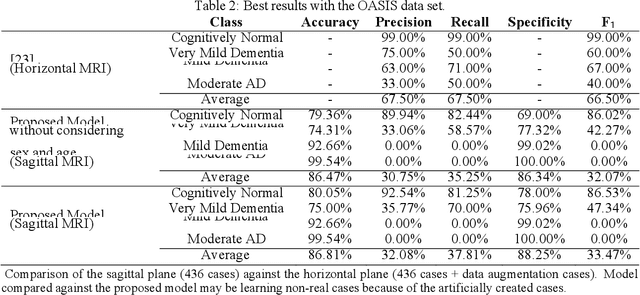
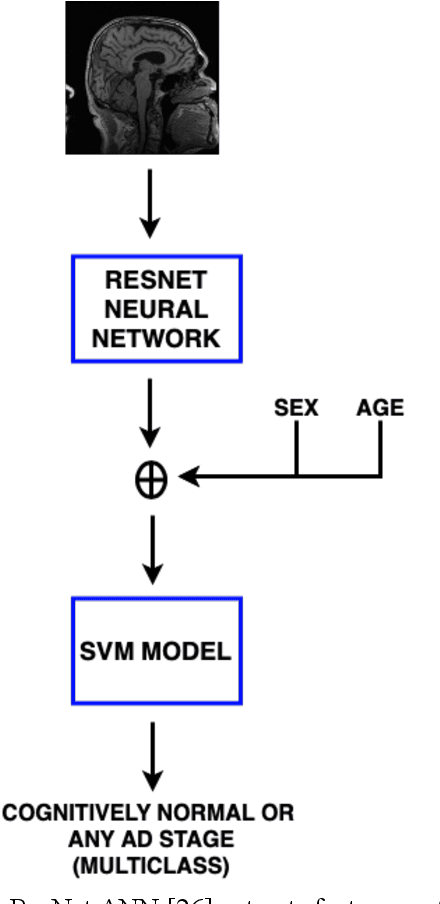
Abstract:Early detection is crucial to prevent the progression of Alzheimer's disease (AD). Thus, specialists can begin preventive treatment as soon as possible. They demand fast and precise assessment in the diagnosis of AD in the earliest and hardest to detect stages. The main objective of this work is to develop a system that automatically detects the presence of the disease in sagittal magnetic resonance images (MRI), which are not generally used. Sagittal MRIs from ADNI and OASIS data sets were employed. Experiments were conducted using Transfer Learning (TL) techniques in order to achieve more accurate results. There are two main conclusions to be drawn from this work: first, the damages related to AD and its stages can be distinguished in sagittal MRI and, second, the results obtained using DL models with sagittal MRIs are similar to the state-of-the-art, which uses the horizontal-plane MRI. Although sagittal-plane MRIs are not commonly used, this work proved that they were, at least, as effective as MRI from other planes at identifying AD in early stages. This could pave the way for further research. Finally, one should bear in mind that in certain fields, obtaining the examples for a data set can be very expensive. This study proved that DL models could be built in these fields, whereas TL is an essential tool for completing the task with fewer examples.
Convolutional Neural Networks for Sleep Stage Scoring on a Two-Channel EEG Signal
Mar 30, 2021
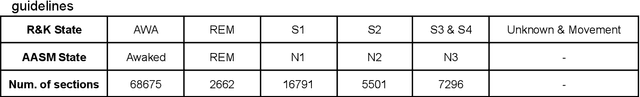
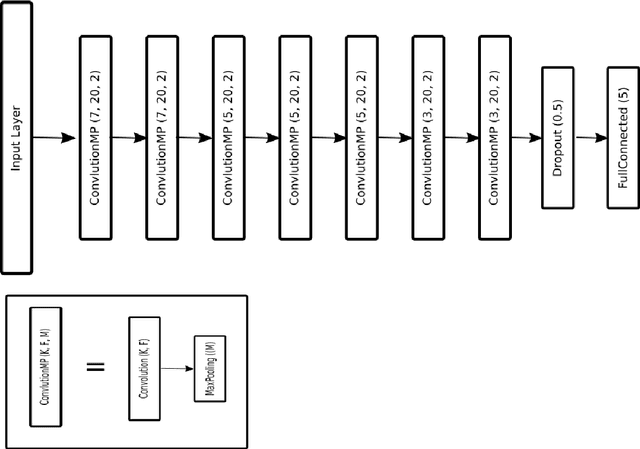
Abstract:Sleeping problems have become one of the major diseases all over the world. To tackle this issue, the basic tool used by specialists is the Polysomnogram, which is a collection of different signals recorded during sleep. After its recording, the specialists have to score the different signals according to one of the standard guidelines. This process is carried out manually, which can be highly time-consuming and very prone to annotation errors. Therefore, over the years, many approaches have been explored in an attempt to support the specialists in this task. In this paper, an approach based on convolutional neural networks is presented, where an in-depth comparison is performed in order to determine the convenience of using more than one signal simultaneously as input. Additionally, the models were also used as parts of an ensemble model to check whether any useful information can be extracted from signal processing a single signal at a time which the dual-signal model cannot identify. Tests have been performed by using a well-known dataset called expanded sleep-EDF, which is the most commonly used dataset as the benchmark for this problem. The tests were carried out with a leave-one-out cross-validation over the patients, which ensures that there is no possible contamination between training and testing. The resulting proposal is a network smaller than previously published ones, but which overcomes the results of any previous models on the same dataset. The best result shows an accuracy of 92.67\% and a Cohen's Kappa value over 0.84 compared to human experts.
* 20 pages, 4 figures, 4 tables
Classical Music Prediction and Composition by means of Variational Autoencoders
Jun 21, 2019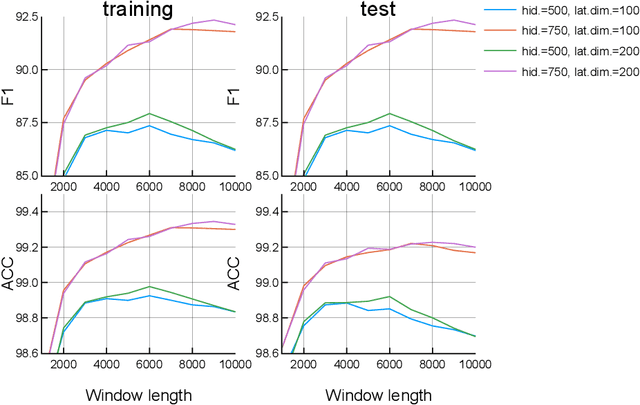
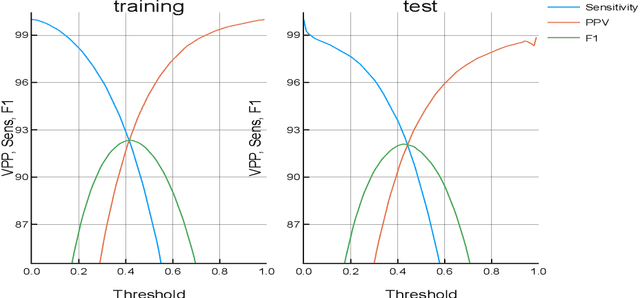
Abstract:This paper proposes a new model for music prediction based on Variational Autoencoders (VAEs). In this work, VAEs are used in a novel way in order to address two different problems: music representation into the latent space, and using this representation to make predictions of the future values of the musical piece. This approach was trained with different songs of a classical composer. As a result, the system can represent the music in the latent space, and make accurate predictions. Therefore, the system can be used to compose new music either from an existing piece or from a random starting point. An additional feature of this system is that a small dataset was used for training. However, results show that the system is able to return accurate representations and predictions in unseen data.
Classification of Two-channel Signals by Means of Genetic Programming
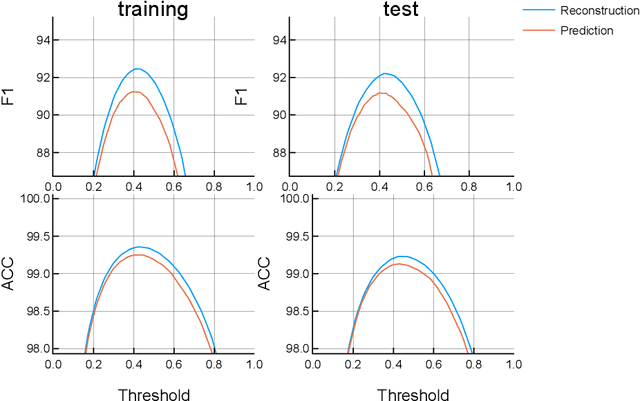
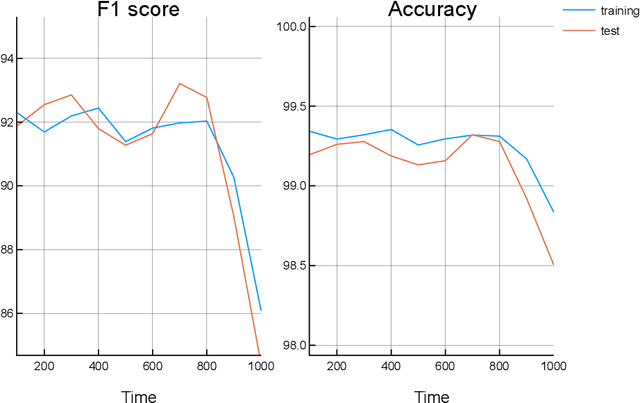
Apr 10, 2019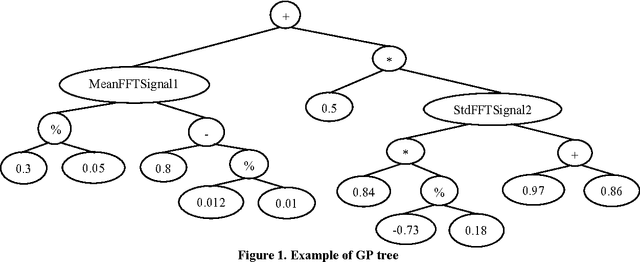
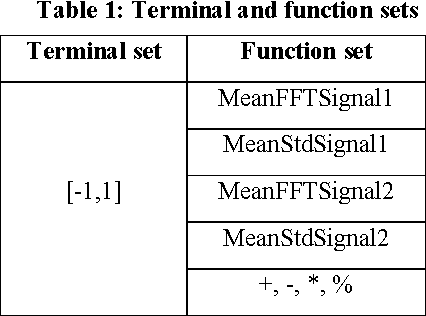
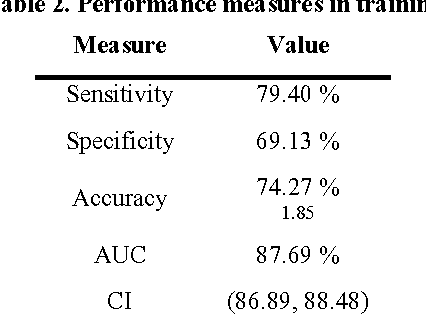
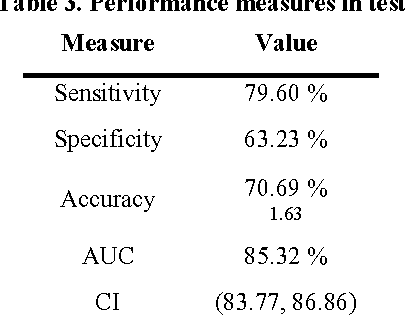
Abstract:Traditionally, signal classification is a process in which previous knowledge of the signals is needed. Human experts decide which features are extracted from the signals, and used as inputs to the classification system. This requirement can make significant unknown information of the signal be missed by the experts and not be included in the features. This paper proposes a new method that automatically analyses the signals and extracts the features without any human participation. Therefore, there is no need for previous knowledge about the signals to be classified. The proposed method is based on Genetic Programming and, in order to test this method, it has been applied to a well-known EEG database related to epilepsy, a disease suffered by millions of people. As the results section shows, high accuracies in classification are obtained
* Conference paper, doble-column, 7 pages, 1 figure, 3 tables
NCBO Ontology Recommender 2.0: An Enhanced Approach for Biomedical Ontology Recommendation
May 25, 2017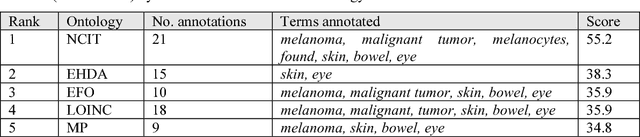
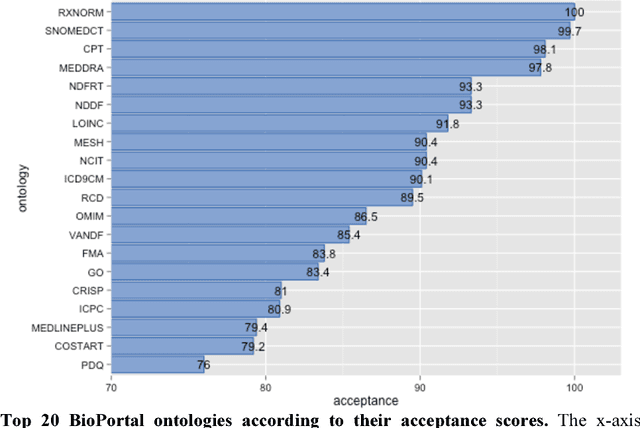
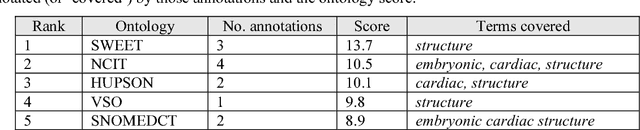
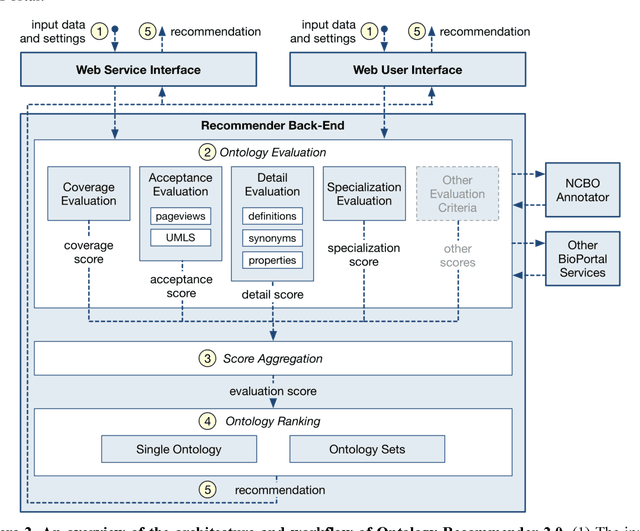
Abstract:Biomedical researchers use ontologies to annotate their data with ontology terms, enabling better data integration and interoperability. However, the number, variety and complexity of current biomedical ontologies make it cumbersome for researchers to determine which ones to reuse for their specific needs. To overcome this problem, in 2010 the National Center for Biomedical Ontology (NCBO) released the Ontology Recommender, which is a service that receives a biomedical text corpus or a list of keywords and suggests ontologies appropriate for referencing the indicated terms. We developed a new version of the NCBO Ontology Recommender. Called Ontology Recommender 2.0, it uses a new recommendation approach that evaluates the relevance of an ontology to biomedical text data according to four criteria: (1) the extent to which the ontology covers the input data; (2) the acceptance of the ontology in the biomedical community; (3) the level of detail of the ontology classes that cover the input data; and (4) the specialization of the ontology to the domain of the input data. Our evaluation shows that the enhanced recommender provides higher quality suggestions than the original approach, providing better coverage of the input data, more detailed information about their concepts, increased specialization for the domain of the input data, and greater acceptance and use in the community. In addition, it provides users with more explanatory information, along with suggestions of not only individual ontologies but also groups of ontologies. It also can be customized to fit the needs of different scenarios. Ontology Recommender 2.0 combines the strengths of its predecessor with a range of adjustments and new features that improve its reliability and usefulness. Ontology Recommender 2.0 recommends over 500 biomedical ontologies from the NCBO BioPortal platform, where it is openly available.
* 29 pages, 8 figures, 11 tables
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge