Carlos Fernandez-Lozano
Random Forest-Based Prediction of Stroke Outcome
Feb 01, 2024Abstract:We research into the clinical, biochemical and neuroimaging factors associated with the outcome of stroke patients to generate a predictive model using machine learning techniques for prediction of mortality and morbidity 3 months after admission. The dataset consisted of patients with ischemic stroke (IS) and non-traumatic intracerebral hemorrhage (ICH) admitted to Stroke Unit of a European Tertiary Hospital prospectively registered. We identified the main variables for machine learning Random Forest (RF), generating a predictive model that can estimate patient mortality/morbidity. In conclusion, machine learning algorithms RF can be effectively used in stroke patients for long-term outcome prediction of mortality and morbidity.
Ensemble of Convolution Neural Networks on Heterogeneous Signals for Sleep Stage Scoring
Jul 23, 2021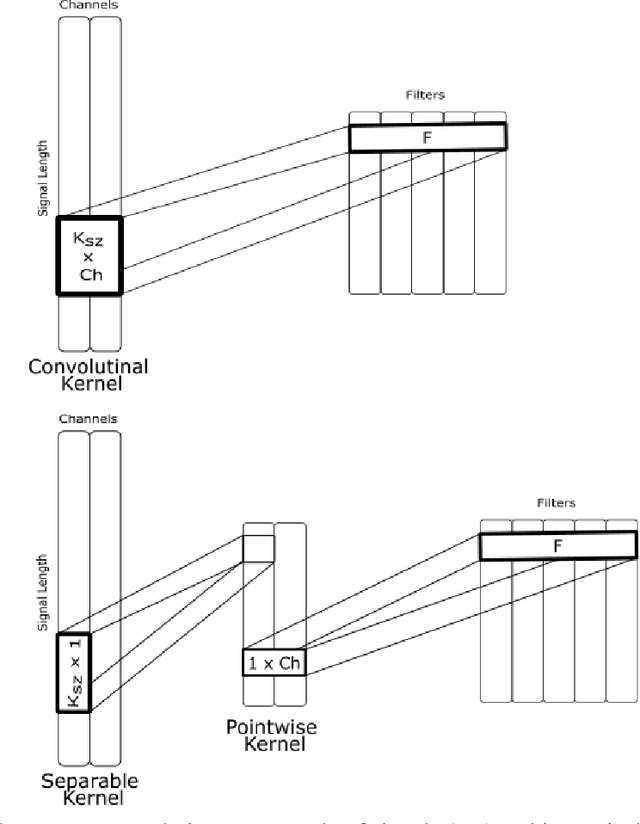

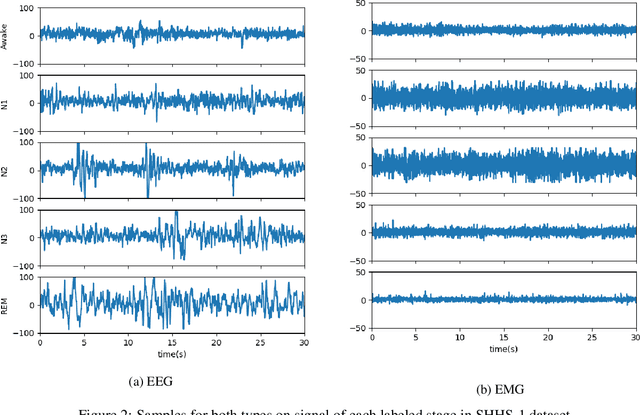
Abstract:Over the years, several approaches have tried to tackle the problem of performing an automatic scoring of the sleeping stages. Although any polysomnography usually collects over a dozen of different signals, this particular problem has been mainly tackled by using only the Electroencephalograms presented in those records. On the other hand, the other recorded signals have been mainly ignored by most works. This paper explores and compares the convenience of using additional signals apart from electroencephalograms. More specifically, this work uses the SHHS-1 dataset with 5,804 patients containing an electromyogram recorded simultaneously as two electroencephalograms. To compare the results, first, the same architecture has been evaluated with different input signals and all their possible combinations. These tests show how, using more than one signal especially if they are from different sources, improves the results of the classification. Additionally, the best models obtained for each combination of one or more signals have been used in ensemble models and, its performance has been compared showing the convenience of using these multi-signal models to improve the classification. The best overall model, an ensemble of Depth-wise Separational Convolutional Neural Networks, has achieved an accuracy of 86.06\% with a Cohen's Kappa of 0.80 and a $F_{1}$ of 0.77. Up to date, those are the best results on the complete dataset and it shows a significant improvement in the precision and recall for the most uncommon class in the dataset.
Classification of signaling proteins based on molecular star graph descriptors using Machine Learning models
Apr 10, 2019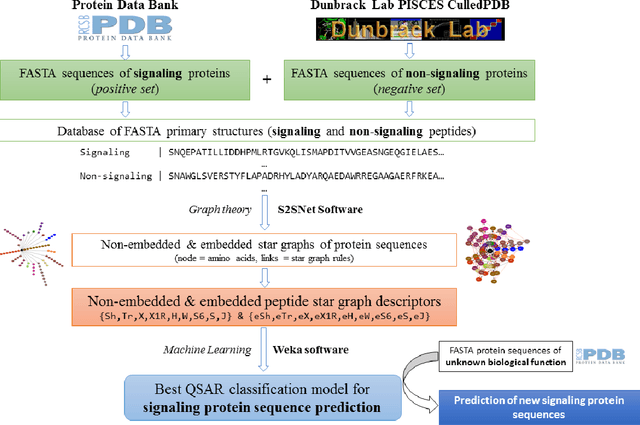
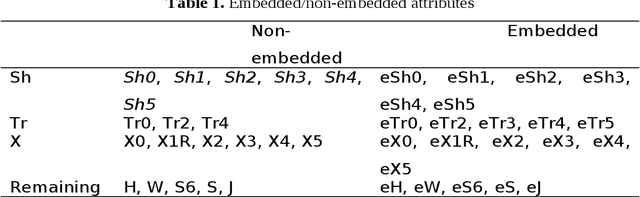
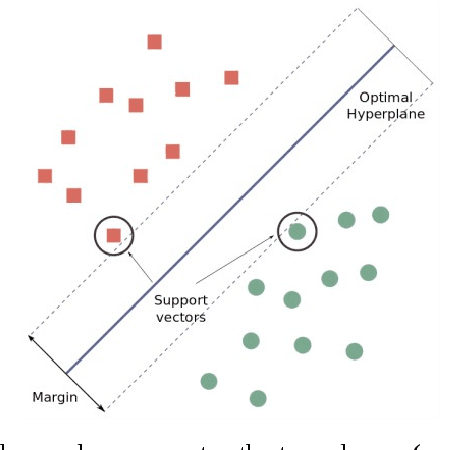
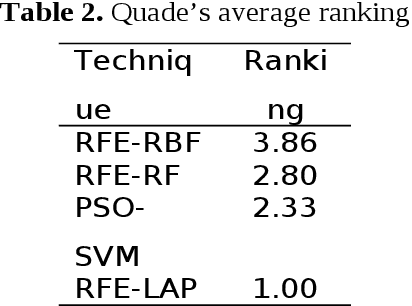
Abstract:Signaling proteins are an important topic in drug development due to the increased importance of finding fast, accurate and cheap methods to evaluate new molecular targets involved in specific diseases. The complexity of the protein structure hinders the direct association of the signaling activity with the molecular structure. Therefore, the proposed solution involves the use of protein star graphs for the peptide sequence information encoding into specific topological indices calculated with S2SNet tool. The Quantitative Structure-Activity Relationship classification model obtained with Machine Learning techniques is able to predict new signaling peptides. The best classification model is the first signaling prediction model, which is based on eleven descriptors and it was obtained using the Support Vector Machines - Recursive Feature Elimination (SVM-RFE) technique with the Laplacian kernel (RFE-LAP) and an AUROC of 0.961. Testing a set of 3114 proteins of unknown function from the PDB database assessed the prediction performance of the model. Important signaling pathways are presented for three UniprotIDs (34 PDBs) with a signaling prediction greater than 98.0%.
* 19 pages, 6 figures, 3 tables
A Hybrid Evolutionary System for Automated Artificial Neural Networks Generation and Simplification in Biomedical Applications
Apr 09, 2019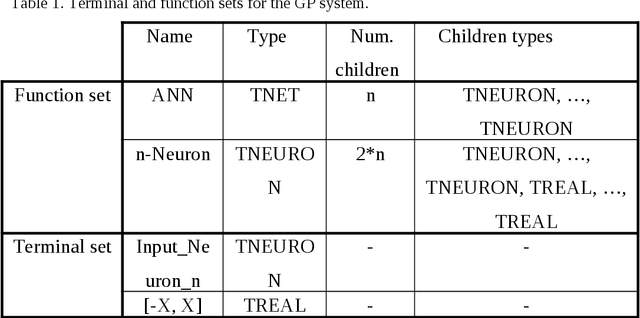
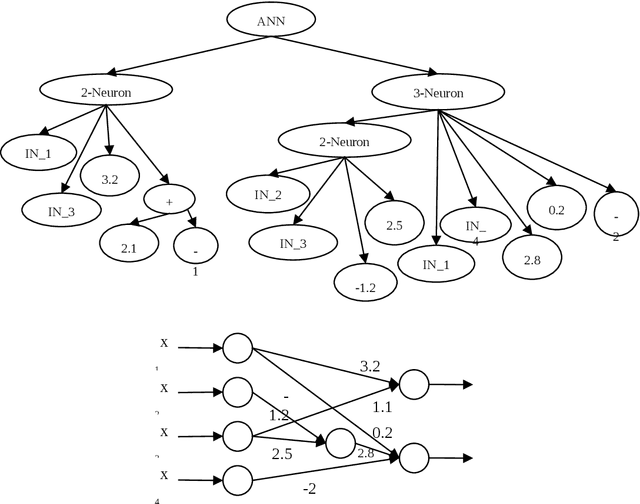
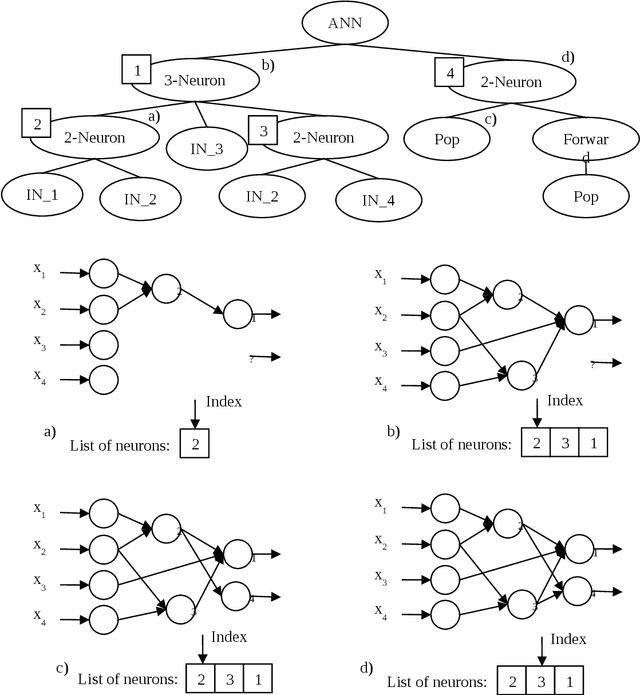
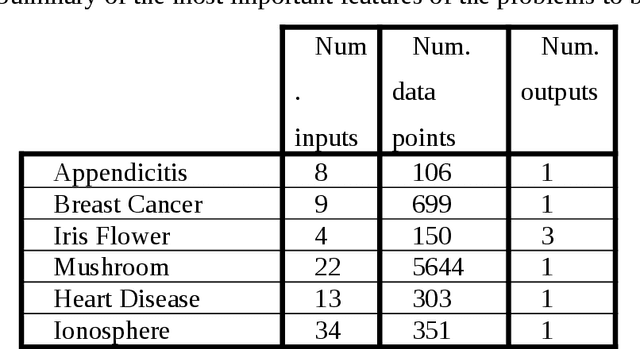
Abstract:Data mining and data classification over biomedical data are two of the most important research fields in computer science. Among the great diversity of techniques that can be used for this purpose, Artifical Neural Networks (ANNs) is one of the most suited. One of the main problems in the development of this technique is the slow performance of the full process. Traditionally, in this development process, human experts are needed to experiment with different architectural procedures until they find the one that presents the correct results for solving a specific problem. However, many studies have emerged in which different ANN developmental techniques, more or less automated, are described. In this paper, the authors have focused on developing a new technique to perform this process over biomedical data. The new technique is described in which two Evolutionary Computation (EC) techniques are mixed to automatically develop ANNs. These techniques are Genetic Algorithms and Genetic Programming. The work goes further, and the system described here allows to obtain simplified networks with a low number of neurons to resolve the problems. The system is compared with the already existent system which also uses EC over a set of well-known problems. The conclusions reached from these comparisons indicate that this new system produces very good results, which in the worst case are at least comparable to existing techniques and in many cases are substantially better.
* 48 pages, 9 figures, 17 tables
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge