Cristian R. Munteanu
Application of Tabular Transformer Architectures for Operating System Fingerprinting
Feb 13, 2025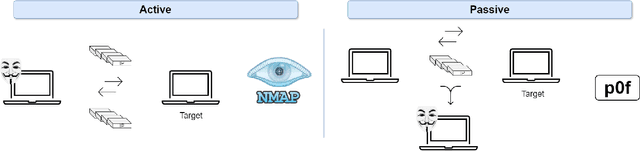
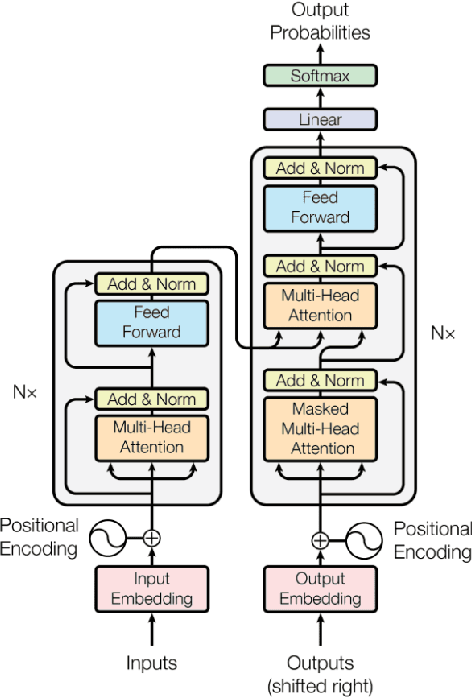

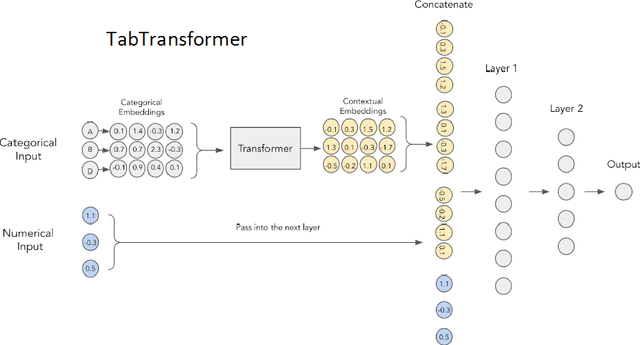
Abstract:Operating System (OS) fingerprinting is essential for network management and cybersecurity, enabling accurate device identification based on network traffic analysis. Traditional rule-based tools such as Nmap and p0f face challenges in dynamic environments due to frequent OS updates and obfuscation techniques. While Machine Learning (ML) approaches have been explored, Deep Learning (DL) models, particularly Transformer architectures, remain unexploited in this domain. This study investigates the application of Tabular Transformer architectures-specifically TabTransformer and FT-Transformer-for OS fingerprinting, leveraging structured network data from three publicly available datasets. Our experiments demonstrate that FT-Transformer generally outperforms traditional ML models, previous approaches and TabTransformer across multiple classification levels (OS family, major, and minor versions). The results establish a strong foundation for DL-based OS fingerprinting, improving accuracy and adaptability in complex network environments. Furthermore, we ensure the reproducibility of our research by providing an open-source implementation.
Automatic Assessment of Alzheimer's Disease Diagnosis Based on Deep Learning Techniques
May 18, 2021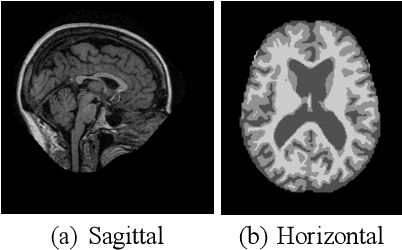
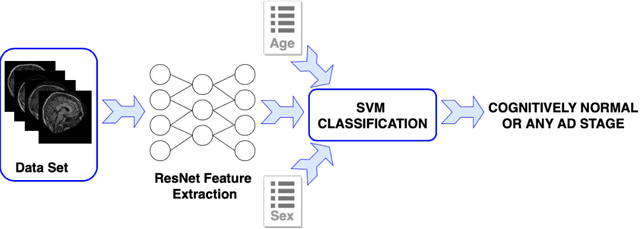
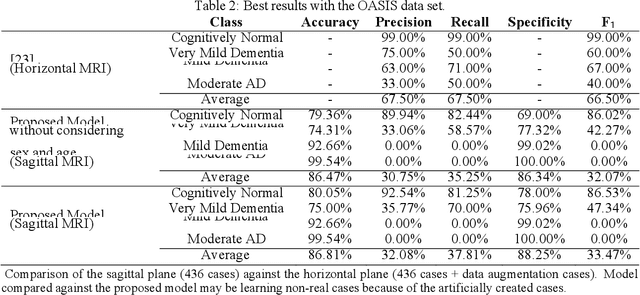
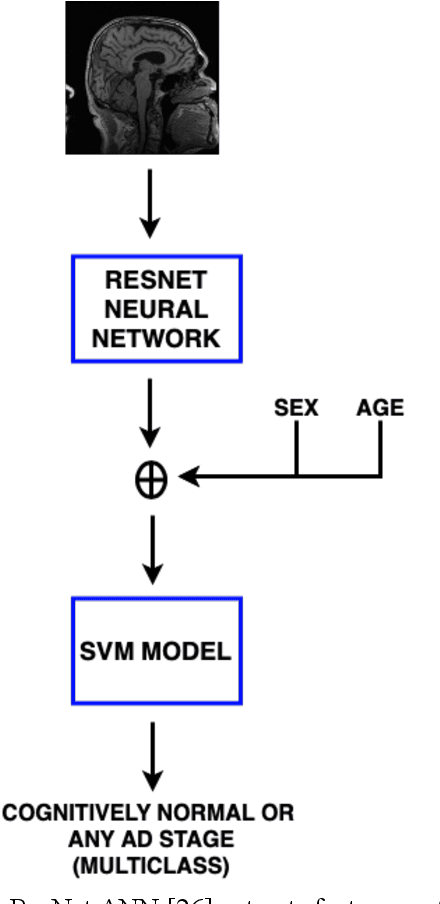
Abstract:Early detection is crucial to prevent the progression of Alzheimer's disease (AD). Thus, specialists can begin preventive treatment as soon as possible. They demand fast and precise assessment in the diagnosis of AD in the earliest and hardest to detect stages. The main objective of this work is to develop a system that automatically detects the presence of the disease in sagittal magnetic resonance images (MRI), which are not generally used. Sagittal MRIs from ADNI and OASIS data sets were employed. Experiments were conducted using Transfer Learning (TL) techniques in order to achieve more accurate results. There are two main conclusions to be drawn from this work: first, the damages related to AD and its stages can be distinguished in sagittal MRI and, second, the results obtained using DL models with sagittal MRIs are similar to the state-of-the-art, which uses the horizontal-plane MRI. Although sagittal-plane MRIs are not commonly used, this work proved that they were, at least, as effective as MRI from other planes at identifying AD in early stages. This could pave the way for further research. Finally, one should bear in mind that in certain fields, obtaining the examples for a data set can be very expensive. This study proved that DL models could be built in these fields, whereas TL is an essential tool for completing the task with fewer examples.
Classification of signaling proteins based on molecular star graph descriptors using Machine Learning models
Apr 10, 2019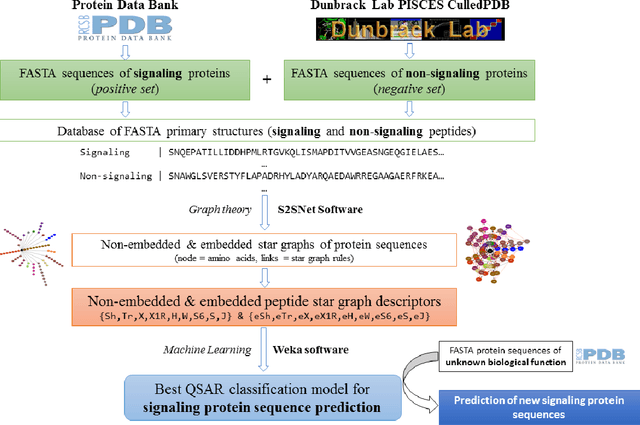
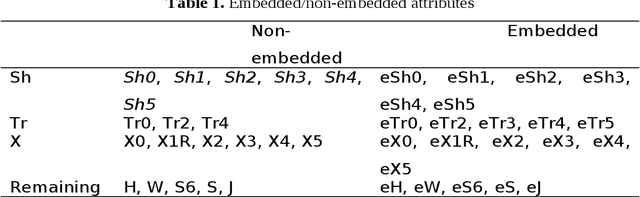
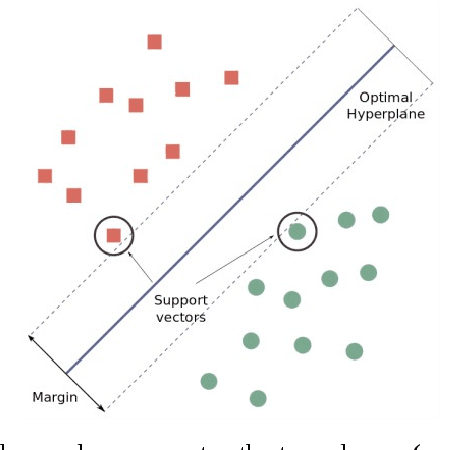
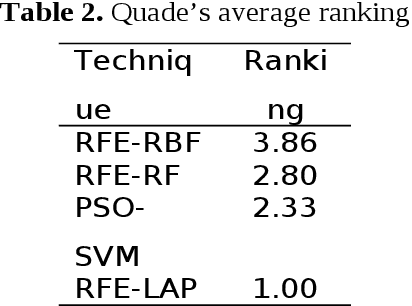
Abstract:Signaling proteins are an important topic in drug development due to the increased importance of finding fast, accurate and cheap methods to evaluate new molecular targets involved in specific diseases. The complexity of the protein structure hinders the direct association of the signaling activity with the molecular structure. Therefore, the proposed solution involves the use of protein star graphs for the peptide sequence information encoding into specific topological indices calculated with S2SNet tool. The Quantitative Structure-Activity Relationship classification model obtained with Machine Learning techniques is able to predict new signaling peptides. The best classification model is the first signaling prediction model, which is based on eleven descriptors and it was obtained using the Support Vector Machines - Recursive Feature Elimination (SVM-RFE) technique with the Laplacian kernel (RFE-LAP) and an AUROC of 0.961. Testing a set of 3114 proteins of unknown function from the PDB database assessed the prediction performance of the model. Important signaling pathways are presented for three UniprotIDs (34 PDBs) with a signaling prediction greater than 98.0%.
* 19 pages, 6 figures, 3 tables
A Hybrid Evolutionary System for Automated Artificial Neural Networks Generation and Simplification in Biomedical Applications
Apr 09, 2019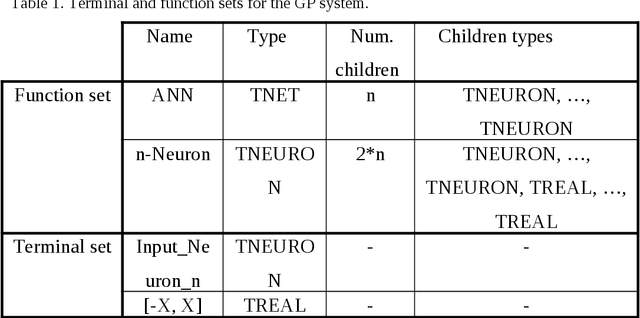
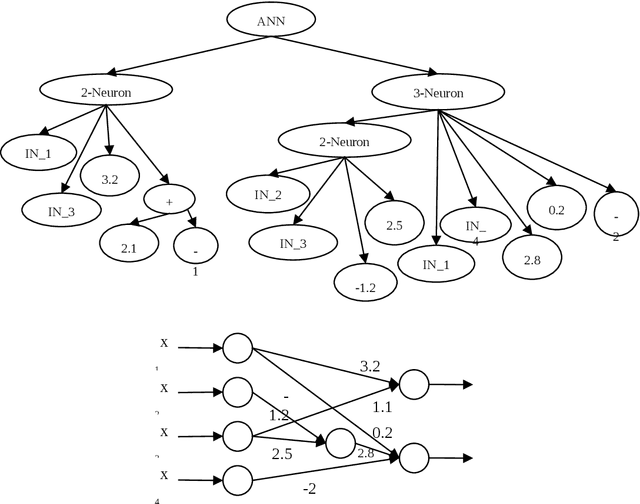
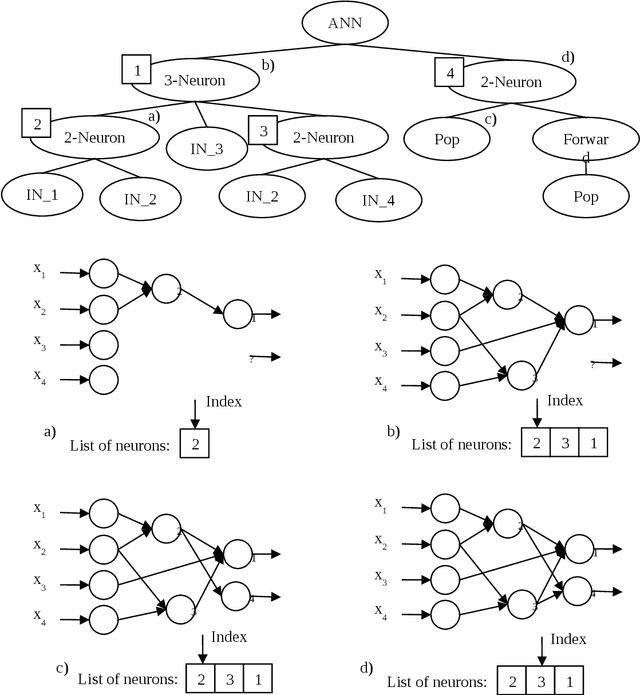
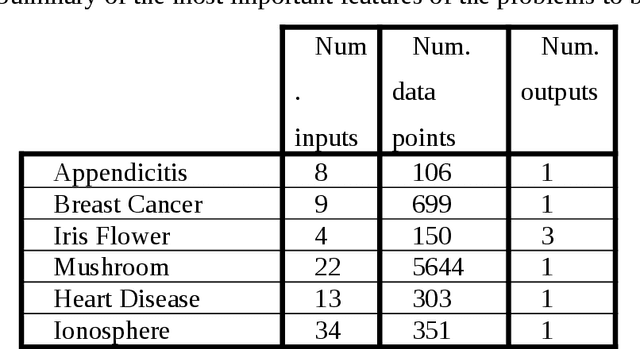
Abstract:Data mining and data classification over biomedical data are two of the most important research fields in computer science. Among the great diversity of techniques that can be used for this purpose, Artifical Neural Networks (ANNs) is one of the most suited. One of the main problems in the development of this technique is the slow performance of the full process. Traditionally, in this development process, human experts are needed to experiment with different architectural procedures until they find the one that presents the correct results for solving a specific problem. However, many studies have emerged in which different ANN developmental techniques, more or less automated, are described. In this paper, the authors have focused on developing a new technique to perform this process over biomedical data. The new technique is described in which two Evolutionary Computation (EC) techniques are mixed to automatically develop ANNs. These techniques are Genetic Algorithms and Genetic Programming. The work goes further, and the system described here allows to obtain simplified networks with a low number of neurons to resolve the problems. The system is compared with the already existent system which also uses EC over a set of well-known problems. The conclusions reached from these comparisons indicate that this new system produces very good results, which in the worst case are at least comparable to existing techniques and in many cases are substantially better.
* 48 pages, 9 figures, 17 tables
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge